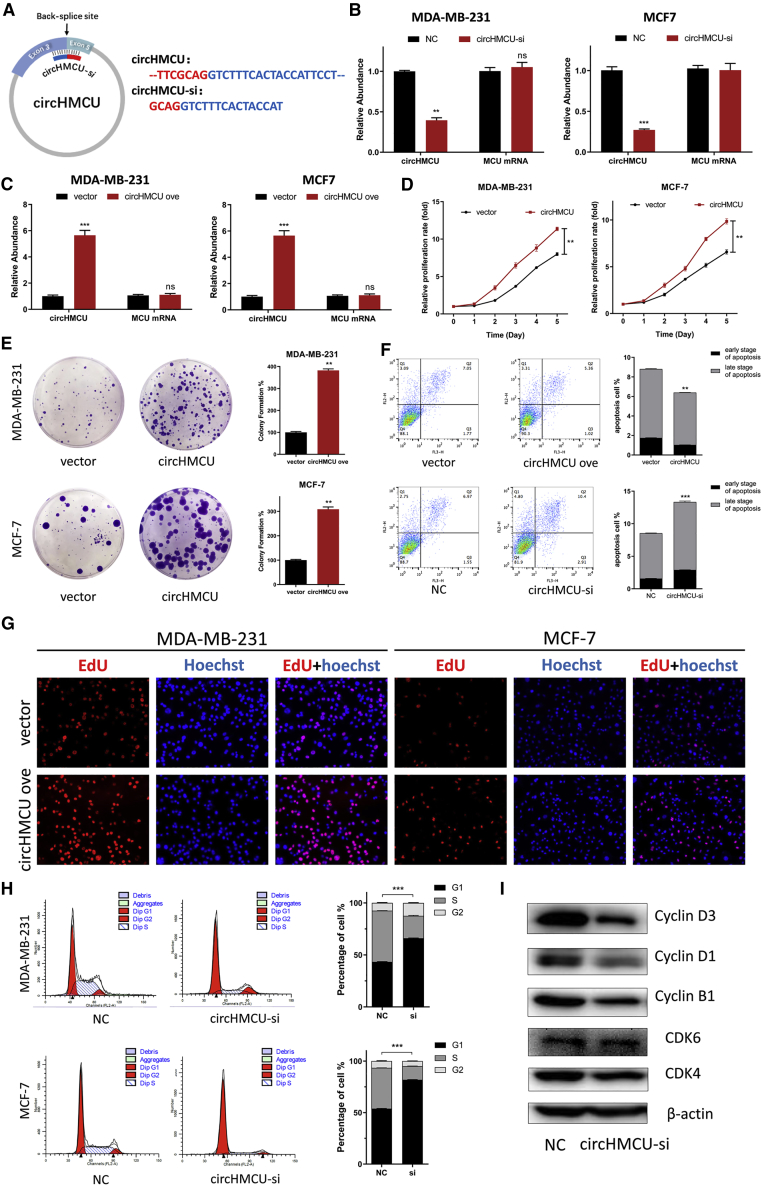
Figure 2.
Effects of circHMCU on Breast Cancer Cell Cycle, Proliferation, and Apoptosis In Vitro
(A) Schematic illustrating the target sequences of the siRNAs specific to the back-splicing junction of circHMCU. (B) The expression levels of circHMCU in MDA-MB-231 cells transfected with NC (negative control) or circHMCU-si were detected by real-time PCR. (C) The expression levels of circHMCU in MDA-MB-231 cells after stable transfection of circHMCU or empty vector plasmids were detected by real-time PCR. (D) MTT assays were used to determine the cell viability of MDA-MB-231 and MDA-MB-468 cells transfected with circHMCU overexpression plasmid. (E) A colony formation assay was performed in MDA-MB-231 and MDA-MB-468 cells stably transfected with circHMCU overexpression plasmid. (F) The overexpression of circHMCU decreased apoptosis in MDA-MB-468 cells. (G) Observation of DNA synthesis of MDA-MB-231 and MDA-MB-468 cells transfected with circHMCU overexpression plasmid by EdU assay. (H) Flow cytometry was performed to determine the effect of circHMCU on changes in cell cycle distribution (left). Statistical diagrams show significant differences (middle). Western blotting was used to detect cyclin B1, cyclin D1, cyclin D3, CDK4, and CDK6 expression (left). Data are the means ± SEM of three experiments. ∗∗p < 0.01, ∗∗∗p < 0.001.