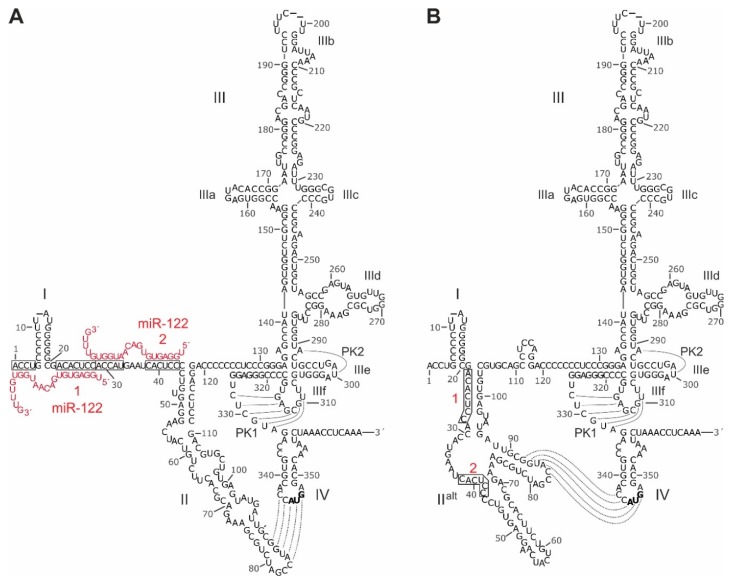
Figure 2.
Sequence and RNA secondary structure of the HCV IRES. (A) The 5′UTR of HCV and its canonical structure [63,109,110,111,112,113,114]. The sequence shown is from genotype 2a (J6 isolate), with length variations among HCV isolates indicated by short horizontal dashes at nucleotide positions. Nucleotide numbering is according to the MAFFT alignment in the supplement of [63] (http://www.rna.uni-jena.de/supplements/hcv/). For comparison, the first nucleotide of the core coding sequence (AUG, in bold) in the SL IV is nucleotide No. 342 in genotype 1b (Con1) and No. 341 in genotype 2a (J6 and JFH1 isolates). IRES SL domains are indicated by roman numerals. microRNA-122 (miR-122, red) binding is indicated, miR-122 target sequences are boxed. Pseudoknot (PK) base pairing and the interaction between SL II and SL IV is indicated; (B) Secondary structure model of the 5′UTR with alternative folding of SL IIalt. The structure is largely according to [72,77,115], with minor modifications according to our RNAalifold outputs using several genotypes (not shown).