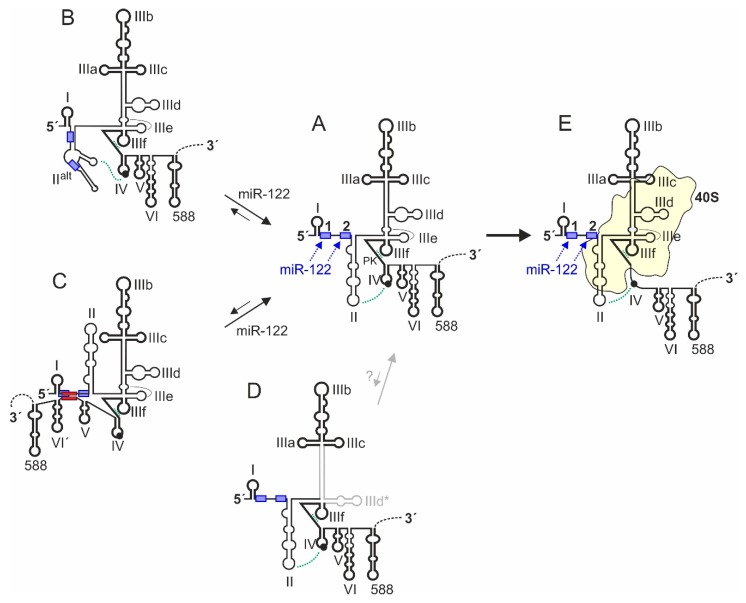
Figure 3.
Conformational dynamics of HCV 5′UTR structure. The conserved canonical 5′UTR structure [63,109,110,111,112,113,114] is shown in the middle (structure A). 5′UTR sequences which have a strong intrinsic tendency to fold into a distinct secondary and tertiary structure are depicted in bold. The constitutive double pseudoknot (PK) interactions involving SL IIIe (PK2) and IIIf (PK1) and the possible interaction between SL II and SL IV are indicated. The 5′UTR structure B [72,77,115] has the region between SLs I and III refolded and forms an alternative version of SL II (SL IIalt). This SL IIalt can form in the absence of miR-122, then hiding both miR-122 binding sites. Binding of miR-122 promotes refolding of structure B to structure A [72,115,122]. The 5′UTR structure C can form by fold-back of nucleotides 428–442 in the base of SL VI in the core protein coding region to hybridize with sequences downstream of SL I. In this state, the translation activity of the IRES is decreased [130,131,132,133,134]. This hybridization also interferes with binding of miR-122 to the 5′UTR. In turn, binding of miR-122 induces refolding of structure C to structure A [135]. Structure D is a yet completely hypothetic structure that was predicted to be the energetically slightly favored conserved structure [63]. In this structure D, the SL IIId of the IRES is refolded to form SL IIId*. However, a possible biological relevance of this hypothetical structure is unclear, in particular since several interactions of the canonical SL IIId have been described (see main text). Structure E is formed upon binding to the ribosomal 40S subunit. Then, SL IV sequences containing the AUG start codon are unfolded and positioned in the mRNA entry channel of the 40S subunit, aided by interaction with SL II.