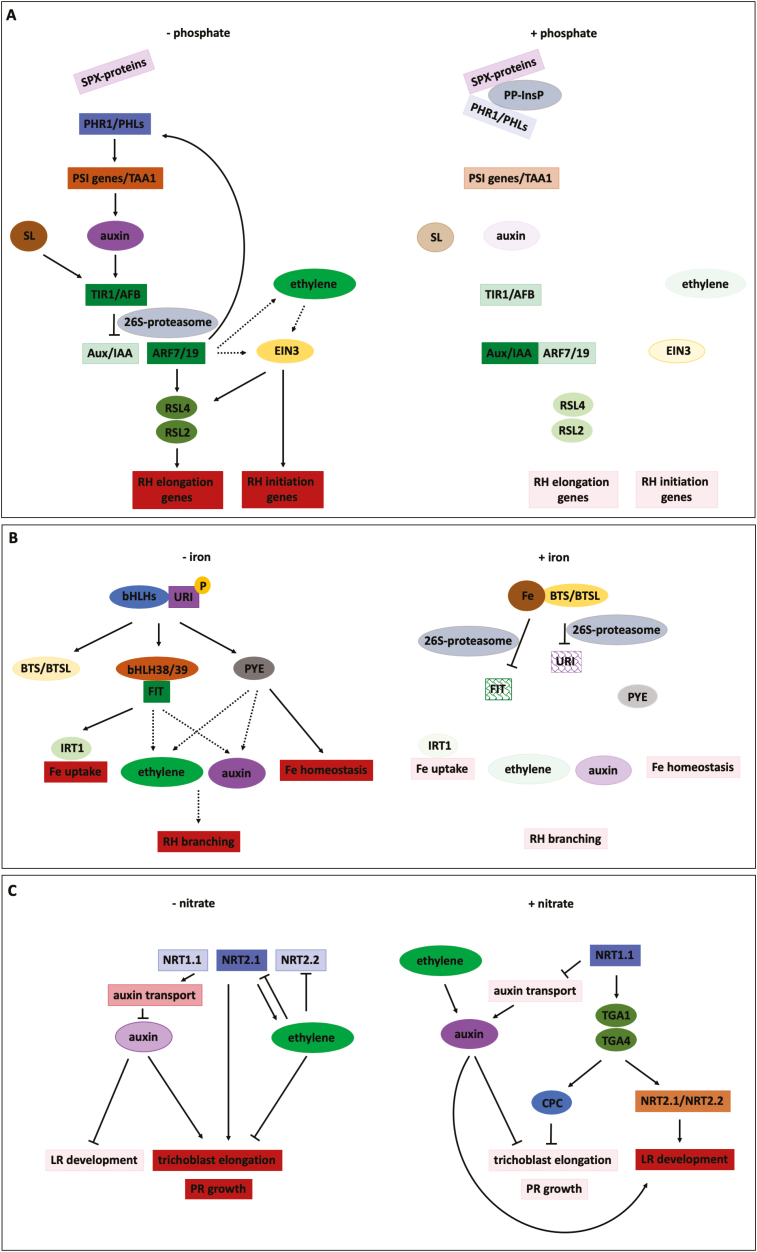
Fig. 5.
Models for signalling pathways downstream of phosphate, iron, and nitrate. Simplified models describing how phosphate (A), iron (B) and nitrate (C) deficiency or presence (left and right, respectively) regulate root hair development. Active proteins are represented in dark colour, inactive proteins are in pale colour, and shingled colour indicates the proteins are targeted for ubiquitination and subsequent degradation by the 26S-proteasome. For hormones, full colour and faded colour represent high and low concentrations. Dotted lines represent unknown relationships/interactions. ARF7/19, AUXIN RESPONSE FACTOR 7/19; Aux/IAA, AUXIN/INDOLE-3-ACETIC ACID; bHLHs, basic helix–loop–helix proteins; BTS, BRUTUS; BTSL, BRUTUS-LIKE; CPC, CAPRICE; EIN3, ETHYLENE INSENSITIVE 3; FIT, FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR; IRT1, IRON-REGULATED TRANSPORTER 1; LR, lateral root; NRT1.1/NRT2.1/NRT2.2, NITRATE TRANSPORTER 1.1/2.1/2.2; PHL, PHOSPHATE STARVATION RESPONSE 1-LIKE; PHR1, PHOSPHATE STARVATION RESPONSE 1; PR, primary root; PSI, phosphate starvation induced; PYE, POPEYE; RH, root hair; RSL2/4, ROOT HAIR DEFECTIVE 6-LIKE 2/4; SL, strigolactone; SPX-proteins, SPX-domain containing proteins; TAA1, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1; TGA1/4, TGACG SEQUENCE-SPECIFIC BINDING PROTEIN 1/4; TIR1/AFB, TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-BOX; URI, UPSTREAM REGULATOR OF IRT 1.