In Enterobacteriales, the AcrAB-TolC efflux pump exports substrates, including antimicrobials, from the cell. Overexpression of AcrAB-TolC can occur after exposure to fluoroquinolones, leading to multidrug resistance. The expression of AcrAB-TolC in Salmonella is primarily regulated by the transcriptional activator RamA. However, other transcriptional activators, such as MarA, SoxRS, and Rob, can influence AcrAB-TolC expression. This study determined whether the overproduction or absence of RamA influences the mutation rate or the phenotype of mutants selected in Salmonella enterica serovar Typhimurium SL1344 after ciprofloxacin exposure.
KEYWORDS: AcrB, RamA, SoxR, multidrug resistance, mutation rate
ABSTRACT
In Enterobacteriales, the AcrAB-TolC efflux pump exports substrates, including antimicrobials, from the cell. Overexpression of AcrAB-TolC can occur after exposure to fluoroquinolones, leading to multidrug resistance. The expression of AcrAB-TolC in Salmonella is primarily regulated by the transcriptional activator RamA. However, other transcriptional activators, such as MarA, SoxRS, and Rob, can influence AcrAB-TolC expression. This study determined whether the overproduction or absence of RamA influences the mutation rate or the phenotype of mutants selected in Salmonella enterica serovar Typhimurium SL1344 after ciprofloxacin exposure. The absence of RamA (SL1344 ramA::aph) resulted in mutation frequencies/rates similar to those of wild-type Salmonella Typhimurium SL1344. However, the overproduction of RamA (SL1344 ramR::aph) and, consequently, AcrB resulted in a significantly higher mutation frequency and rate than for wild-type Salmonella Typhimurium SL1344. Whole-genome sequencing revealed that in addition to selecting gyrA mutants resistant to quinolones, SL1344 and SL1344 ramA::aph also produced multidrug-resistant (MDR) mutants, associated with mutations in soxR. Conversely, mutations in SL1344 ramR::aph occurred in gyrA only. Although transcriptional regulators such as SoxRS are believed to play a minor role in AcrAB-TolC regulation under antibiotic selective pressure, we show that soxR mutants can be selected after exposure to ciprofloxacin, including when RamA is absent. This demonstrates that under selective pressure, Salmonella can respond to increased efflux pump expression by mutating other AcrAB-TolC regulatory genes, allowing for the evolution of MDR. Understanding how Salmonella responds to antibiotic pressure in the absence/overproduction of RamA is important if targeting transcriptional regulators to alter efflux is to be considered an avenue for future drug discovery.
INTRODUCTION
Antimicrobial resistance is one of the great global challenges facing modern medicine (1). Bacteria can be intrinsically resistant to certain antibiotics but can evolve via chromosomal mutation and can also acquire resistance by horizontal transfer of resistance genes. Mutations that result in antimicrobial resistance typically alter antibiotic activity by one of the following mechanisms: modification of the drug target, reduced membrane permeability, and increased efflux. Those mutations that reduce the intracellular accumulation of antibiotics by increasing efflux confer reduced susceptibility to a range of different antimicrobial classes and can cause multidrug resistance (MDR). Therefore, extensive research surrounding the development of compounds capable of neutralizing this resistance mechanism has been undertaken.
Listed by the World Health Organization (WHO) as a high-priority pathogen for which new treatments are urgently needed, fluoroquinolone-resistant Salmonella enterica causes a significant health burden worldwide (2). Resistance upon exposure to the fluoroquinolone ciprofloxacin frequently results from mutations in the topoisomerase-encoding genes gyrA, gyrB, parC, and parE. However, MDR resulting from ciprofloxacin exposure can occur in Gram-negative bacteria as a result of the overproduction of efflux pumps (3, 4). In Salmonella enterica, increased active efflux is attributed mainly to the overexpression of the AcrAB-TolC efflux pump (5). Although subject to multiple levels of regulation, in Salmonella, AcrAB-TolC is primarily regulated by RamA, an AraC/XylS transcriptional activator (6). When ramA is highly expressed, there is a concomitant overexpression of acrAB and tolC, which results in the increased translation of the AcrAB-TolC pump proteins, leading to MDR (Fig. 1) (7). In the absence of RamA, it is difficult to select MDR mutants (8).
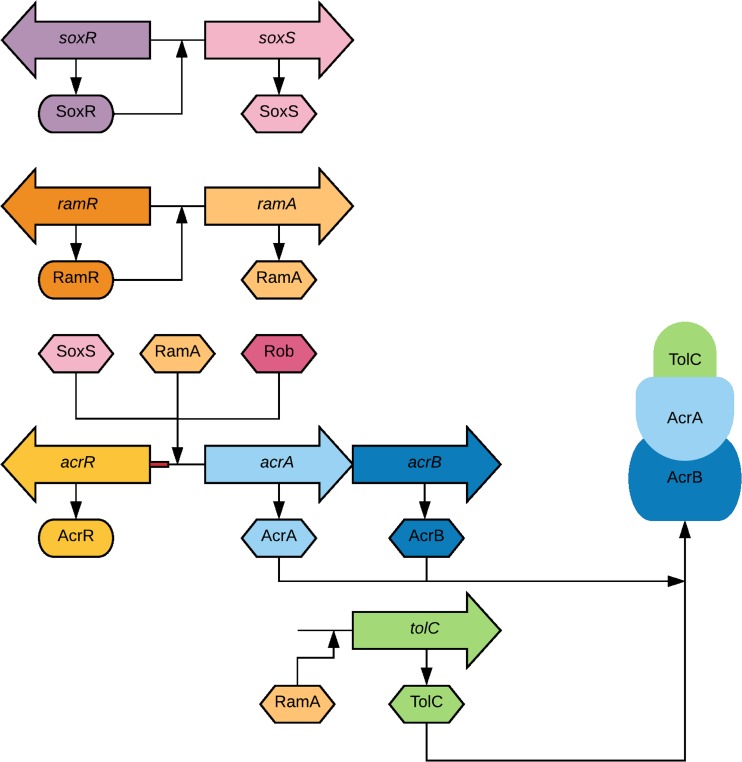
FIG 1.
Schematic representation of the known regulatory pathways for expression of the AcrAB-TolC efflux pump in Salmonella. The genes are represented as arrows, and their translated proteins are represented as ovals (transcriptional repressors) and hexagons (transcriptional activators). The AcrAB-TolC pump extrudes drugs across the cytoplasmic and outer membranes. Excessive production of AcrA and AcrB is prevented by the local repressor AcrR. Activation of acrA, acrB, and tolC transcription occurs primarily due to the global regulatory protein RamA by binding to the rambox upstream of these genes. As demonstrated, the regulatory proteins SoxS and Rob can also activate acrAB-tolC transcription. RamA expression is controlled by RamR, which represses the activation of ramA. Likewise, SoxR controls the expression of both soxR and soxS.
In addition to RamA, in Enterobacteriales, the transcriptional activators MarA, SoxRS, and Rob are also capable of regulating the expression of AcrAB-TolC (Fig. 1) (5). Although mutations that increase ramA expression are often reported in clinical and veterinary isolates of Salmonella and Escherichia coli, MDR due to mutations within transcriptional regulators such as soxR has been observed (9–12). The soxRS regulatory locus is responsible for the response of Enterobacteriales to oxidative stress. In the absence of a stressor, the [2Fe-2S] iron clusters of SoxR are reduced, and the protein is inactive. Upon oxidative stress, the iron clusters are oxidized, and SoxR is able to stimulate the transcription of soxS (13). SoxS, like RamA, is part of the AraC family of transcriptional activators (5, 14, 15). When activated, SoxS is able to cause an increase in the transcription of all of the genes within its regulon; this includes acrAB-tolC (13). In the absence of AcrB, soxS expression increases, probably as a response to increased oxidative stress caused by the lack of this major efflux pump (14). This suggests that there are feedback mechanisms by which Enterobacteriales use different transcriptional regulators to maintain efflux.
Capable of increasing bacterial susceptibility to currently available antimicrobials, inhibition of efflux pumps is an important potential avenue to tackle MDR (16). Targeting transcriptional regulators, such as RamA in Salmonella, may reduce the ability of the organism to develop MDR via overexpression of AcrAB-TolC. Understanding how Salmonella responds to selective pressure in the absence or overproduction of RamA is important. Furthermore, knowing if, in the presence of an AcrAB-TolC substrate, the bacterium is capable of acquiring mutations allowing it to circumnavigate inhibition via RamA-regulated pathways, is important when considering the use of transcriptional regulators as drug targets and to improve our understanding of the regulation of multidrug efflux. Antibiotic selective pressure can trigger a plethora of cellular events, which can determine the phenotype of any resultant mutant that evolves during drug exposure; whether this occurs early or late within the growth of a population may affect the mutation rate. Given that bacteria with higher acrAB expression levels have lower expression levels of the DNA mismatch repair gene mutS, lower growth rates, and higher mutation frequencies, selective pressure that leads to increased expression of the AcrAB-TolC system may contribute to increased mutation rates (17).
In this study, we set out to determine whether different levels of ramA expression result in differences in mutation rates and the mechanism by which resistance to the fluoroquinolone ciprofloxacin evolves.
RESULTS
The rate and frequency of mutation upon exposure to ciprofloxacin are dependent on the level of expression of the transcriptional activator RamA.
When SL1344 was exposed to ciprofloxacin at the MIC, the average frequency of mutation (proportion of mutant cells in a population) was 3.82 × 10−8 mutations/cell/generation; the average rate of mutation (rate at which mutation events arise) was 4.08 × 10−9 mutations/cell/generation (Table 1 and Fig. 2 and 3). At the MIC of ciprofloxacin for SL1344 ramA::aph, the frequency of mutation was similar to that for the wild type; the mutation frequency and rate were 7.15 × 10−8 and 5.11 × 10−9 mutations/cell/generation, respectively. Interestingly, SL1344 ramR::aph, which overexpresses ramA and leads to the concomitant overexpression of acrAB, had a significantly higher mutation frequency and rate than wild-type SL1344, at 2.54 × 10−7 and 3.03 × 10−8 mutations/cell/generation, respectively.
TABLE 1.
Average frequencies and rates of mutation (per cell/per generation) of S. Typhimurium wild-type SL1344 and the SL1344 ramR::aph and SL1344 ramA::aph mutants upon exposure to MICs of ciprofloxacina
| Strain | Genotype | Ciprofloxacin MIC (μg/ml) | Mean frequency ± SD | Mean mutation rate per cell/per generation ± SD |
|---|---|---|---|---|
| SL1344 | Wild type | 0.03 | 3.82 × 10−8 ± 9.75 × 10−9 | 4.08 × 10−9 ± 1.15 × 10−9 |
| L1322 | ramA::aph | 0.03 | 7.15 × 10−8 ± 2.47 × 10−8 | 5.11 × 10−9 ± 9.75 × 10−10 |
| L1007 | ramR::aph | 0.12 | 2.54 × 10−7 ± 9.52 × 10−8 | 3.03 × 10−8 ± 9.21 × 10−9 |
The MIC of each parental strain is shown.
FIG 2.
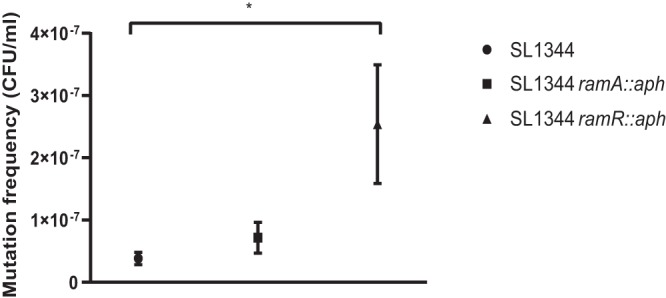
Mutation frequencies of strains exposed to ciprofloxacin. The mutation frequency was calculated as the average total number of ciprofloxacin-resistant colonies divided by the viable count. *, P < 0.05, calculated using one-way ANOVA relative to wild-type SL1344 (n = 30 independent replicates).
FIG 3.
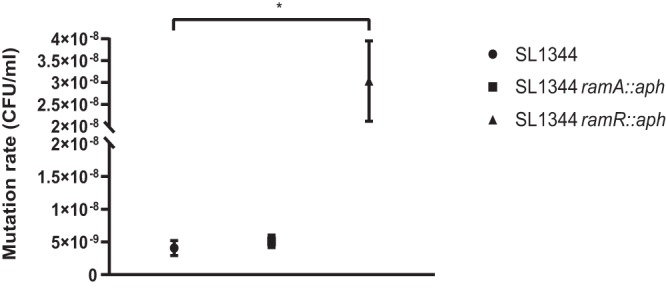
Mutation rates of strains exposed to ciprofloxacin. The phenotypic mutation rate, μ, was calculated using the Lea Coulson method of the median. *, P < 0.05, calculated using one-way ANOVA relative to wild-type SL1344 (n = 30 independent replicates).
Unless RamA is already overexpressed, ciprofloxacin selects for MDR mutants.
When SL1344 was exposed to ciprofloxacin, MDR mutants with decreased susceptibility to ciprofloxacin, nalidixic acid, chloramphenicol, and ampicillin were obtained (Table 2). Whole-genome sequencing (WGS) of one representative, L1881, revealed a SNP (single-nucleotide polymorphism) conferring a missense mutation (D137N) in the transcriptional repressor soxR in which aspartic acid was substituted for asparagine. In contrast to the wild-type strain, mutants selected from SL1344 ramR::aph were not MDR but had reduced susceptibility to both ciprofloxacin and nalidixic acid, a result of a substitution of aspartic acid for glycine within the quinolone resistance-determining region (QRDR) of GyrA. Prior to WGS, all mutants were passaged on antibiotic-free media; the mutations identified were confirmed by PCR and subsequent DNA sequencing of the amplimers.
TABLE 2.
MIC phenotypes of the mutants obtained after exposure to ciprofloxacina
| S. Typhimurium strain | Genotype | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|---|
| CIP | NAL | CHL | TET | EtBr | AMP | ||
| SL1344 | WT | 0.03 | 8 | 4 | 1 | 1,024 | 1 |
| L1881 | SoxR Asp137Asn | 0.12 | 16 | 16 | 2 | >2,048 | 8 |
| SL1344 | ramA::aph | 0.03 | 8 | 4 | 1 | 1,024 | 1 |
| L1886 | GyrA Ser83Phe | 0.5 | >64 | 4 | 1 | 2,048 | 2 |
| L1882 | GyrA Asp87Gly | 0.12 | >64 | 4 | 1 | 1,024 | 2 |
| L1880 | SoxR Asn134Thr | 0.12 | 16 | 16 | 2 | 2,048 | 8 |
| SL1344 | ramR::aph | 0.12 | 16 | 16 | 4 | 2,048 | 8 |
| L1877 | GyrA Asp87Gly | 0.5 | >64 | 16 | 4 | 2,048 | 8 |
The genotypes determined by WGS of each mutant are shown. Italic values indicate a ≥4-fold increase in the MIC in comparison to that for the parental strain. Boldface type indicates SoxR mutants. WT, wild type; CIP, ciprofloxacin; NAL, nalidixic acid; CHL, chloramphenicol; TET, tetracycline; EtBr, ethidium bromide; AMP, ampicillin.
When RamA is absent, ciprofloxacin can still select MDR mutants.
In the absence of RamA (SL1344 ramA::aph), exposure to ciprofloxacin gave rise to two phenotypically different mutants: those that were MDR and those that were resistant to quinolones only. One mutant, L1880, had decreased susceptibility to ciprofloxacin, nalidixic acid, chloramphenicol, and ampicillin. WGS revealed a SNP conferring a missense mutation in soxR with a substitution of asparagine for threonine at position 134 (N134T). Mutants resistant to only quinolones possessed gyrA mutations conferring Ser83Phe or Asp87Gly substitutions (mutants L1886 and L1882).
DISCUSSION
As described by ourselves and others, when ciprofloxacin was used as a selecting agent, both (fluoro)quinolone-resistant and MDR mutants were obtained from wild-type Salmonella (3, 18, 19). Antibiotic treatment fluctuation assays were performed at the MIC, as mutation selection experiments at sub-MICs are likely to alter the mutation rate and phenotype of the mutants selected (20).
The estimated frequency of mutation for S. Typhimurium after exposure to ciprofloxacin at the MIC is reported to be ∼10−9, which is in the range reported in this study (8, 21). The mutation frequency will measure all the mutants present in a population at a given time, irrespective of whether the mutation event occurred early or late during the growth of that population. Calculating mutation rates can be very complex but aims for a more accurate frequency of mutational events in a population in the presence of an antibiotic and is important in predicting the emergence of antibiotic-resistant bacteria under a particular selective pressure. The rate of mutation shown here was also in keeping with that reported in a previous study (22).
It has been well documented that gyrA mutations at codons 83 and 87 confer ciprofloxacin resistance (23). Selecting bacteria with mutations that interfere with binding to the target site of quinolones is not an unexpected mechanism by which Salmonella strains can develop resistance to ciprofloxacin. Mutations that confer MDR typically give rise to “low-level” resistance to a broad spectrum of antibiotics, and target site mutations are also necessary to provide high-level resistance (24). Therefore, when ciprofloxacin is present and able to interact with its intracellular target, even at very low concentrations (as is the case when RamA is overexpressed), selective pressure is exerted that drives the evolution of target site mutations in gyrA. Therefore, gyrA mutants are likely to occur in both the absence and overexpression of efflux pumps.
Given that in S. Typhimurium, RamA is the primary regulator of acrAB-tolC transcription, it was interesting to find that the MDR seen for the SL1344 mutant (L1881) did not result from a mutation in ramA; rather, a mutation in soxR was observed. soxR is typically upregulated in response to oxidative stress, leading to an increased expression of soxS (5, 14). This mutational event is very uncommon but has been described for clinical isolates of Salmonella and E. coli; these soxR mutations were associated with resistance to fluoroquinolones and chloramphenicol (9–12). In the MDR mutant (L1880) selected from SL1344 (ramA::aph), a soxR mutation was also found. Zheng et al. demonstrated that ramA inactivation caused an altered transcription of genes regulated by soxS, suggesting coregulation between ramA and soxS (25). When acrB is deleted, soxS expression increases; it is hypothesized that this is a response to increased oxidative stress caused by the lack of activity of this major efflux pump (14). It is likely, therefore, that in the absence of RamA, mutations enabling the increased production of SoxS are selected in order to maintain functional efflux and allow for bacterial survival.
The crystal structure of SoxR from E. coli revealed that each monomer consists of an N-terminal DNA-binding domain, a dimerization helix domain, and a C-terminal domain with a [2Fe-2S] cluster (26–28). The [2Fe-2S] cluster is vital for SoxR to function, and it is stabilized by α3′- and α5′-helices (26, 27). These areas are highly conserved among all Enterobacteriaceae, including Salmonella (29). The mutations at positions 134 and 137 in the two mutants (L1880 and L1881) lie very close to the α5′-helix and the conserved cysteines for [2Fe-2S] cluster binding (30). Mutations in Salmonella and E. coli within neighboring regions have been shown to alter redox potential and consequently create conformational changes that interfere with the DNA-binding domain of SoxR (9, 27, 29). We hypothesize that the missense mutations described in the two mutants will have similar effects in Salmonella, enabling the mutant to overexpress SoxS and result in MDR.
In response to ciprofloxacin exposure, MDR mutants have been shown to occur as a result of RamA overproduction (3). However, in mutant selection experiments using SL1344 (ramR::aph) that overexpressed RamA, none of the mutants contained mutations that confer additional increased transcription of efflux pumps or caused MDR. These results suggest that further mutations in efflux-regulatory genes would not create an additional fitness advantage. This hypothesis is supported by evidence from clinical isolates demonstrating that the fitness costs of a mutation impact the nature of subsequent second-step mutations, in preference to the mutation rate alone (30). It is hypothesized that second-step mutations conferring additional increased transcription of an efflux pump would confer a high fitness cost (30). This may also explain why the majority of mutants from SL1344 (ramA::aph) had mutations in gyrA as opposed to mutations altering efflux pump gene regulation.
We have shown that natural overexpression of acrAB via the lack of RamR repression of RamA in Salmonella enterica affects the mutation rate and frequency. This is in keeping with results obtained when artificial levels of acrAB are produced. El Meouche and Dunlop noted that plasmid-mediated overexpression of acrAB resulted in a higher mutation frequency than for wild-type E. coli and S. Typhimurium LT2 (17). Here, we show that chromosomally mediated overexpression of AcrAB, via deletion of the transcriptional repressor RamR (ramR::aph), resulted in a higher rate of mutation and a higher frequency of mutation than in cells with wild-type AcrAB levels; deletion of ramR as a means to overexpress acrAB was chosen as the experimental strategy as the levels of AcrB produced are more likely to reflect those observed in a clinical isolate. Overexpression of acrAB in E. coli results in a mutator phenotype because of lower expression levels of the DNA mismatch repair gene mutS (17). This deficiency in mutS expression results in an inability to repair the misincorporation of bases that occurs during replication (31). Overexpression of stress response mechanisms, including efflux pumps, can incur a fitness cost by increasing cellular energy requirements and by the removal of metabolites that are essential for bacterial growth (32). We hypothesize that the mutator phenotype occurs in order to compensate for fitness costs that may result from the overexpression of acrAB.
After exposure to ciprofloxacin, mutations in soxR can confer MDR in S. enterica serovar Typhimurium in both the presence and absence of RamA. When ramA is already overexpressed, further mutations in the genes encoding transcriptional regulators of the AcrAB-TolC pump did not occur. SoxRS is traditionally believed to play a minor role in the regulation of AcrAB-TolC in Salmonella; however, in response to antimicrobial selective pressure, mutations in the transcriptional regulator soxR can confer a survival advantage and confer MDR in the presence of normal and impaired regulation of the AcrAB-TolC efflux pump. Inhibition of regulatory genes of AcrAB-TolC, including ramA and marA, is postulated to be a method to reduce antibiotic resistance by keeping efflux levels low, thereby increasing the intracellular concentration of antibiotics and increasing their activity. However, we show that in the absence of RamA, compensatory mutations appear within soxR that result in MDR. This is important when considering the usefulness of compounds that behave as efflux inhibitors. Future studies evaluating novel approaches to tackling antibiotic resistance by targeting efflux in Enterobacteriales, including Salmonella, such as inhibition of transcription factors, will need to consider all adaptive responses when designing future experiments.
MATERIALS AND METHODS
Bacterial strains and mutant selection.
S. enterica serovar Typhimurium strain SL1344 and its mutants with deletions in RamA (SL1344 ramA::aph) or RamR (SL1344 ramR::aph) were used throughout. SL1344 ramA::aph and ramR::aph were constructed previously by Ricci et al. (3, 8). Reverse transcription-PCR experiments performed previously determined that the expression levels of ramA were undetectable for SL1344 ramA::aph and were increased 25-fold for SL1344 ramR::aph relative to wild-type SL1344 (33). Bacteria were routinely cultured in Lennox broth unless otherwise indicated.
Spontaneous mutants with decreased susceptibility to fluoroquinolones were selected using a fluctuation assay (34). Thirty independent cultures for each parental strain were grown aerobically at 37°C for 16 to 20 h in antibiotic-free Lennox broth, concentrated by centrifugation, and resuspended in sterile Lennox broth to give a cell density of approximately 109 CFU/ml. Using a spiral plater (Don Whitely Scientific, UK), 50 μl of each suspension was used to inoculate a Lennox broth agar plate containing the MIC of ciprofloxacin for each strain and incubated aerobically at 37°C for up to 3 days (Table 1). To calculate viable counts, each culture grown overnight was diluted to 104 CFU/ml and 105 CFU/ml; 50 μl of each dilution was sufficient to enable single-colony identification, enabling viable counts to be calculated. Each mutant selection experiment was repeated on three separate occasions.
Calculation of the mutation frequency and rate of mutations.
The mutation frequency was calculated as the average total number of ciprofloxacin-resistant colonies divided by the viable count. The phenotypic mutation rate, μ, was calculated using the Lea Coulson method of the median (34, 35). The equations [r/m − ln(m) − 1.24] = 0 and μ = m/2Nt were used, where r is the average number of colonies obtained, m is the number of mutants per culture obtained, and Nt is the final number of cells in culture (35). One-way analysis of variance (ANOVA) was used to determine statistical differences in mutation frequencies and rates between wild-type S. Typhimurium SL1344 and SL1344 ramR::aph or SL1344 ramA::aph.
Susceptibility to antibiotics.
Ten colonies from each fluctuation assay were randomly selected to determine the phenotypes of putative mutants. All antibiotics and dyes were obtained from Sigma (Poole, UK). The susceptibilities of putative mutants to six AcrAB-TolC substrates (ciprofloxacin, nalidixic acid, chloramphenicol, tetracycline, ampicillin, and ethidium bromide) were determined (36). The MIC of each agent was determined by the standardized agar doubling-dilution method as described by the British Society of Antimicrobial Chemotherapy (BSAC) (37). For ciprofloxacin, a cutoff value of 0.25 mg/liter was used to define resistance (8, 38, 39). Mutants were classed as MDR if there was a 2-fold decrease in susceptibility to at least three classes of antimicrobials compared to the parent strain (8).
Whole-genome sequencing and PCR.
One mutant of each phenotype (as determined by susceptibility testing) was whole-genome sequenced (WGS). Genomic DNA was extracted using a bacterial genomic DNA isolation kit (Norgen BioTek Corporation) according to the manufacturer’s instructions. Paired-end sequencing was carried out by the Beijing Genomics Institute (BGI) (Hong Kong, China) using the Illumina HiSeq 4000 platform. Raw sequences were assessed for quality with FASTQC. The sequencing depth was 60×. Comparisons were made with the genome of the SL1344 strain from the Ensembl database (ASM21085v2) using SNIPPY to determine any single-nucleotide polymorphisms (SNPs). Alignment was performed using bowtie2. BAM files were created and compared using Artemis (Sanger Institute, UK) to confirm any SNPs detected using SNIPPY. The minimum coverage to call an SNP was 10 with a confidence cutoff value of 0.9. Where any SNPs were identified, the amino acid sequence was compared using Clustal Omega to identify whether the SNP correlated with a missense mutation and the corresponding protein change.
PCR and DNA sequencing were performed to confirm SNPs within genes of interest. The primers used are shown in Table 3. DNA sequencing of PCR amplimers was carried out at the Functional Genomics Laboratory (University of Birmingham, UK).
TABLE 3.
Primers used in this study to confirm SNPs identified by WGS
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| gyrA | CGTTGGTGACGTAATCGGTA | CCGTACCGTCATAGTTATCC |
| soxR | CAATGTTTAGCGGTTGGTCG | AATCATCTTCAAGCAGCCGG |
ACKNOWLEDGMENTS
We thank Robert Marshall and Xuan Wang-Kan for helpful discussions and critical appraisal of the manuscript.
This work was supported by a research grant from the MRC (MR/P022596/1). Elizabeth M. Grimsey was supported by an MRC iCASE studentship (MR/N017846/1).
REFERENCES
- 1.World Health Organization. 2015. Global action plan on antimicrobial resistance WHO, Geneva, Switzerland. [Google Scholar]
- 2.Cuypers WL, Jacobs J, Wong V, Klemm EJ, Deborggraeve S, Van Puyvelde S. 2018. Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb Genom 4:e000195. doi: 10.1099/mgen.0.000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricci V, Piddock LJ. 2009. Ciprofloxacin selects for multidrug resistance in Salmonella enterica serovar Typhimurium mediated by at least two different pathways. J Antimicrob Chemother 63:909–916. doi: 10.1093/jac/dkp054. [DOI] [PubMed] [Google Scholar]
- 4.Poole K. 2000. Efflux-mediated resistance to fluoroquinolones in Gram-negative bacteria. Antimicrob Agents Chemother 44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weston N, Sharma P, Ricci V, Piddock LJV. 2018. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res Microbiol 169:425–431. doi: 10.1016/j.resmic.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Abouzeed YM, Baucheron S, Cloeckaert A. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 52:2428–2434. doi: 10.1128/AAC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishino K, Yamaguchi A. 2004. Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J Bacteriol 186:1423–1429. doi: 10.1128/jb.186.5.1423-1429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricci V, Tzakas P, Buckley A, Piddock LJ. 2006. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob Agents Chemother 50:38–42. doi: 10.1128/AAC.50.1.38-42.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koutsolioutsou A, Martins EA, White DG, Levy SB, Demple B. 2001. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob Agents Chemother 45:38–43. doi: 10.1128/AAC.45.1.38-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oethinger M, Podglajen I, Kern WV, Levy SB. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother 42:2089–2094. doi: 10.1128/AAC.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kehrenberg C, Cloeckaert A, Klein G, Schwarz S. 2009. Decreased fluoroquinolone susceptibility in mutants of Salmonella serovars other than Typhimurium: detection of novel mutations involved in modulated expression of ramA and soxS. J Antimicrob Chemother 64:1175–1180. doi: 10.1093/jac/dkp347. [DOI] [PubMed] [Google Scholar]
- 12.O’Regan E, Quinn T, Pagès J-M, McCusker M, Piddock LJV, Fanning S. 2009. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar Enteritidis: involvement of RamA and other global regulators. Antimicrob Agents Chemother 53:1080–1087. doi: 10.1128/AAC.01005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hidalgo E, Leautaud V, Demple B. 1998. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J 17:2629–2636. doi: 10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaves DJ, Ricci V, Piddock LJ. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob Agents Chemother 48:1145–1150. doi: 10.1128/aac.48.4.1145-1150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RG, Gillette WK, Rosner JL. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol Microbiol 35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- 16.Piddock LJV. 2014. Understanding the basis of antibiotic resistance: a platform for drug discovery. Microbiology 160:2366–2373. doi: 10.1099/mic.0.082412-0. [DOI] [PubMed] [Google Scholar]
- 17.El Meouche I, Dunlop MJ. 2018. Heterogeneity in efflux pump expression predisposes antibiotic-resistant cells to mutation. Science 362:686–690. doi: 10.1126/science.aar7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 44:1223–1228. doi: 10.1128/aac.44.5.1223-1228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Dai M, Hao H, Wang Y, Huang L, Almofti YA, Liu Z, Yuan Z. 2011. The role of RamA on the development of ciprofloxacin resistance in Salmonella enterica serovar Typhimurium. PLoS One 6:e23471. doi: 10.1371/journal.pone.0023471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez JL, Baquero F. 2000. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother 44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webber MA, Randall LP, Cooles S, Woodward MJ, Piddock LJ. 2008. Triclosan resistance in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 62:83–91. doi: 10.1093/jac/dkn137. [DOI] [PubMed] [Google Scholar]
- 22.Ricci V, Loman N, Pallen M, Ivens A, Fookes M, Langridge GC, Wain J, Piddock LJV. 2012. The TCA cycle is not required for selection or survival of multidrug-resistant Salmonella. J Antimicrob Chemother 67:589–599. doi: 10.1093/jac/dkr515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooper DC. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis 7:337–341. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R, Swick MC, Ledesma KR, Yang Z, Hu M, Zechiedrich L, Tam VH. 2012. Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother 56:1680–1685. doi: 10.1128/AAC.05693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng J, Tian F, Cui S, Song J, Zhao S, Brown EW, Meng J. 2011. Differential gene expression by RamA in ciprofloxacin-resistant Salmonella Typhimurium. PLoS One 6:e22161. doi: 10.1371/journal.pone.0022161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe S, Kita A, Kobayashi K, Miki K. 2008. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci U S A 105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Weiss B. 1991. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol 173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee KL, Singh AK, Heo L, Seok C, Roe JH. 2015. Factors affecting redox potential and differential sensitivity of SoxR to redox-active compounds. Mol Microbiol 97:808–821. doi: 10.1111/mmi.13068. [DOI] [PubMed] [Google Scholar]
- 29.Huseby DL, Pietsch F, Brandis G, Garoff L, Tegehall A, Hughes D. 2017. Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol Biol Evol 34:1029–1039. doi: 10.1093/molbev/msx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes D, Andersson DI. 2017. Evolutionary trajectories to antibiotic resistance. Annu Rev Microbiol 8:579–596. doi: 10.1146/annurev-micro-090816-093813. [DOI] [PubMed] [Google Scholar]
- 31.Li B, Tsui HC, LeClerc JE, Dey M, Winkler ME, Cebula TA. 2003. Molecular analysis of mutS expression and mutation in natural isolates of pathogenic Escherichia coli. Microbiology 149:1323–1331. doi: 10.1099/mic.0.26213-0. [DOI] [PubMed] [Google Scholar]
- 32.Du Toit A. 2017. Efflux pumps, fitness and virulence. Nat Rev Microbiol 15:512–513. doi: 10.1038/nrmicro.2017.97. [DOI] [PubMed] [Google Scholar]
- 33.Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J, Piddock LJV. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol 192:1607–1616. doi: 10.1128/JB.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pope CF, O’Sullivan DM, McHugh TD, Gillespie SH. 2008. A practical guide to measuring mutation rates in antibiotic resistance. Antimicrob Agents Chemother 52:1209–1214. doi: 10.1128/AAC.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster PL. 2006. Methods for determining spontaneous mutation rates. Methods Enzymol 409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blair JMA, Piddock LJV. 2016. How to measure export via bacterial multidrug resistance efflux pumps. mBio 7:e00840-16. doi: 10.1128/mBio.00840-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews JM, Howe RA, BSAC Working Party on Susceptibility Testing . 2011. BSAC standardized disc susceptibility testing method (version 10). J Antimicrob Chemother 66:2726–2757. doi: 10.1093/jac/dkr359. [DOI] [PubMed] [Google Scholar]
- 38.Aarestrup FM, Wiuff C, Molbak K, Threlfall EJ. 2003. Is it time to change fluoroquinolone breakpoints for Salmonella spp.? Antimicrob Agents Chemother 47:827–829. doi: 10.1128/aac.47.2.827-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wain J, Hoa NT, Chinh NT, Vinh H, Everett MJ, Diep TS, Day NP, Solomon T, White NJ, Piddock LJV, Parry CM. 1997. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis 25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]