The pandemic of coronavirus diseases 2019 (COVID-19) imposes a heavy burden on medical resources [1]. Whether there is correlation between viral load and disease severity has not been clarified. In the study, we retrospectively collected the virological data, as well as demographic, epidemiological clinical information of 92 patients with confirmed COVID-19 in a single hospital in Zhejiang Province, China. We compared the baseline viral loads between severe patients and those mild to moderate at admission and also between those developing severe disease during hospitalization and those not.
We studied 92 patients with confirmed COVID-19 who were admitted from January 19, 2020, to March 19, 2020, in the First Affiliated Hospital of Zhejiang University. The sputum specimens were collected from the lower respiratory tract of each patient at admission and the levels of viral nuclei acid were determined by a real-time PCR (RT-PCR) approach and indicated by the cycle threshold (Ct) values of RT-PCR assays [2]. Other demographic, epidemiological and clinical information were collected and inputted into a pre-designated electronic data collection form. All patients followed up to March 15, 2020. All the statistical analyses were performed using GraphPad Prism 5 (GraphPad Software Inc.; San Diego, CA, USA) and SPSS 20.0 (SPSS Inc.; Chicago, IL, USA).
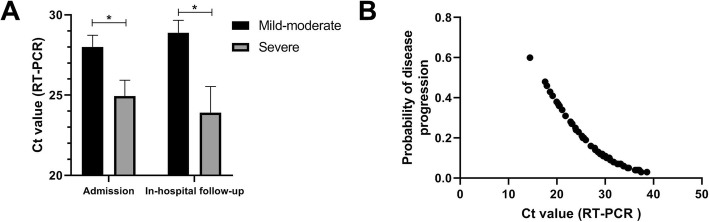
Of the 92 patients, 30 were severe on admission. Of the other 62 mild-moderate cases at admission, 11 cases became severe during hospitalization. The demographic, epidemiological and clinical information was shown in Table 1. All patients were tested for SARS-CoV-2 nucleic acid on sputum specimens from the lower respiratory tract at admission. As shown in Fig. 1a, severe patients had significantly lower Ct values than mild-moderate cases at admission (25 vs. 28, p = 0.017), suggesting a higher viral load in the lower respiratory tract. Furthermore, a higher viral load was observed in sputum specimens from patients who became severe during the hospitalization than those did not (24 vs. 29, p = 0.008). As shown in Fig. 1b, the Ct values of RT-PCR assays negatively correlated with the probability of progression to severe type in all the patients representing mild-to-moderate at admission.
Table 1.
Demographic, comorbidities, epidemiological characteristics, and clinical and laboratory findings of patients with confirmed COVID-19 at admission
| Variables | Total (n = 92) | Mild-moderate at admission | Severe at admission (n = 30) | P value* | |||
|---|---|---|---|---|---|---|---|
| Persistent mild-moderate during hospitalization (n = 51) | Mild-moderate to severe during hospitalization (n = 11) | P value# | Total (n = 62) | ||||
| Demographic data | |||||||
| Age (years) | 55 ± 16 | 49 ± 13 | 59 ± 17 | 0.032 | 51 ± 15 | 63 ± 16 | 0.001 |
| Sex | |||||||
| Male | 57 (62%) | 26 (51%) | 8 (72.7%) | 34 (54.8%) | 23 (76.7%) | ||
| Female | 35 (38%) | 25 (49%) | 3 (27.3%) | 0.189 | 28 (45.2%) | 7 (23.3%) | 0.043 |
| Occupation | |||||||
| Agricultural worker | 45 (48.9%) | 25 (49%) | 7 (63.6%) | 32 (51.6%) | 13 (43.3%) | ||
| Self-employed | 21 (22.8%) | 15 (29.4%) | 2 (18.7%) | 17 (27.4%) | 4 (13.3%) | ||
| Employee | 8 (8.7%) | 5 (9.8%) | 0 (0%) | 5 (8.1%) | 3 (10%) | ||
| Retired | 17 (18.5%) | 5 (9.8%) | 2 (18.2%) | 7 (11.3%) | 10 (33.3%) | ||
| Students | 1 (1.1%) | 1 (2%) | 0 (0%) | 0.669 | 1 (1.6%) | 0 (0%) | 0.082 |
| Smoking history | |||||||
| Yes | 16 (17.4%) | 7 (13.7%) | 3 (27.3%) | 17 (27.4%) | 6 (20%) | ||
| No | 76 (82.6%) | 44 (86.3%) | 8 (72.7%) | 0.268 | 45 (72.6%) | 24 (80%) | 0.441 |
| Comorbidities | |||||||
| Hypertension | 33 (35.9%) | 10 (19.6%) | 7 (63.6%) | 0.003 | 17 (27.4%) | 16 (53.3%) | 0.016 |
| Diabetes | 9 (9.8%) | 1 (2%) | 2 (18.2%) | 0.024 | 3 (4.8%) | 6 (20%) | 0.022 |
| Cardiovascular disease | 8 (8.7%) | 2 (3.9%) | 1 (9.1%) | 0.472 | 3 (4.8%) | 5 (16.7%) | 0.060 |
| Chronic liver diseases | 4 (4.3%) | 2 (3.9%) | 1 (9.1%) | 0.472 | 3 (4.8%) | 1 (3.3%) | 0.741 |
| Chronic renal diseases | 3 (3.3%) | 0 (0%) | 1 (9.1%) | 0.031 | 1 (1.6%) | 2 (6.7%) | 0.203 |
| Others | 6 (6.5%) | 0 (0%) | 2 (18.2%) | 0.002 | 2 (3.2%) | 4 (13.3%) | 0.067 |
| Epidemiological characteristics | |||||||
| Exposure to confirmed cases | 46 (50%) | 30 (58.8%) | 5 (45.5%) | 0.421 | 35 (56.5%) | 11 (36.7%) | 0.077 |
| Family cluster | 27 (29.3%) | 15 (29.4%) | 4 (36.4%) | 0.653 | 19 (30.6%) | 8 (26.7%) | 0.696 |
| Recent travel or residence to/in epidemic area | 25 (27.2%) | 11 (21.6%) | 4 (36.4%) | 0.303 | 15 (24.2%) | 10 (33.3%) | 0.358 |
| Signs and symptoms | |||||||
| Fever | 84 (91.3%) | 45 (88.2%) | 11 (100%) | 0.235 | 56 (90.3%) | 28 (93.3%) | 0.633 |
| Cough | 58 (63%) | 32 (62.7%) | 7 (63.6%) | 0.956 | 39 (62.9%) | 13 (43.3%) | 0.968 |
| Fatigue | 6 (6.5%) | 1 (2%) | 2 (18.2%) | 0.024 | 3 (4.8%) | 3 (10%) | 0.350 |
| Diarrhea | 7 (7.6%) | 1 (2%) | 1 (9.1%) | 0.229 | 2 (3.2%) | 5 (16.7%) | 0.023 |
| Nausea and vomiting | 4 (4.3%) | 3 (5.9%) | 1 (9.1%) | 0.697 | 4 (6.5%) | 0 (0%) | 0.157 |
| Shortness of breath | 25 (27.2%) | 2 (3.9%) | 4 (36.4%) | 0.001 | 6 (9.7%) | 19 (63.3%) | < 0.001 |
| Time to admission | 3 (4) | 4 (3) | 1 (4) | 0.011 | 4 (4) | 1 (4) | 0.211 |
| Time to confirmed diagnosis | 5 (5) | 5 (4) | 4 (4) | 0.160 | 5 (4) | 3 (6) | 0.239 |
| Laboratory parameters | |||||||
| WBC | 6.5 (5.9) | 5.2 (4.1) | 7.5 ± 3.4 | 0.188 | 5.4 (4.5) | 10.8 ± 5.6 | < 0.001 |
| Lymphocyte | 0.8 (0.6) | 0.97 ± 0.47 | 0.7 (0.4) | 0.147 | 0.9 (0.7) | 0.5 (0.5) | 0.001 |
| Platelet | 191 (76) | 193 (83) | 170 ± 56 | 0.159 | 192 (84) | 191 ± 45 | 0.851 |
| CRP | 27 (37) | 13 (27) | 37 ± 27 | 0.036 | 16 (30) | 39 (29) | < 0.001 |
| ALT | 23 (22) | 23 (24) | 17 (15) | 0.338 | 22 (23) | 23 (16) | 0.723 |
| AST | 22 (16) | 21 (12) | 21 (18) | 1.000 | 21 (12) | 26 (23) | 0.236 |
| Cr | 75 (25) | 71 ± 26 | 84 (39) | 0.054 | 73 (28) | 84 (33) | 0.019 |
| INR | 0.98 (0.09) | 0.97 ± 0.08 | 0.97 (0.04) | 0.507 | 0.97 ± 0.06 | 1.01 ± 0.09 | 0.050 |
| Bilirubin | 10.8 (6.0) | 12.2 (5.0) | 10.0 (6.0) | 0.912 | 10.6 (5.0) | 12.6 (9.0) | 0.097 |
| LDH | 281 ± 105 | 227 (103) | 279 ± 101 | 0.376 | 229 (113) | 339 (121) | < 0.001 |
| CK | 70 (76) | 63 (61) | 76 (60) | 0.495 | 64 (58) | 97 (172) | 0.011 |
| Urea nitrogen | 5.3 (3.7) | 4.4 (1.7) | 6.8 (6.9) | < 0.001 | 4.6 (2.2) | 7.7 (4.2) | < 0.001 |
| CT scan | |||||||
| Normal | 3 (3.3%) | 3 (5.9%) | 0 (0%) | 3 (4.8%) | 0 (0%) | ||
| Local lesion | 5 (5.4%) | 4 (7.8%) | 0 (0%) | 4 (6.5%) | 1 (3.3%) | ||
| Multi-lesions | 84 (91.3%) | 44 (86.3%) | 11 (100%) | 1.000 | 55 (88.7%) | 29 (96.7%) | 1.000 |
| ICU admission | 27 (29.3%) | 0 (0%) | 8 (72.7%) | < 0.001 | 8 (12.9%) | 19 (63.3%) | < 0.001 |
Data are expressed as number (percent), mean ± standard deviation (SD), or median (IQR)
#P values comparing data between patients becoming severe and those who did not during hospitalization by the Mann-Whitney U test, chi-squared test, or Fisher’s exact test
*P values comparing data between mild-moderate patients and severe patients at admission
Fig. 1.
a Comparison of baseline sputum viral load between severe and mild-to-moderate patients at admission or between those becoming severe and those did not during the hospitalization. b Relationship between the estimated probability of disease progression during the hospitalization and baseline sputum viral load. Viral load is indicated by the Ct value of RT-PCR assay. The asterisk indicates a P value < 0.05
We found that the viral load of the sputum specimen in the lower respiratory tract tested at baseline is closely related to the severity of COVID-19. More importantly, patients with a higher baseline viral load are more likely to become severe. This finding apparently justifies the concept that early antiviral treatment, if effective, would reduce the risk of progression and thereby the mortality, which has been demonstrated in influenza [3]. In our study, sputum specimens were used, instead of nasopharyngeal and oropharyngeal swabs because it has been shown that samples from lower respiratory tract generally contain a higher level of viral load than nasopharyngeal and oropharyngeal swabs [4] and acquiring swabs is uncomfortable for patients.
In summary, we found a positive association between sputum viral load and disease severity as well as risk of progression.
Acknowledgements
Not applicable.
Authors’ contributions
J.S conceptualized the idea and designed the study. X.Y and S.S drafted the manuscript and J.S revised it. X.Y, S.S, Y.S, H.W, and R.Z participated in the data collection, analysis, and interpretation. The authors read and approved the final manuscript.
Funding
This work was supported by grants from Chinese National Natural science foundation (no. 81670567 and 81870425) and the Fundamental Research Funds for the Central Universities.
Availability of data and materials
The datasets and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study is reviewed and approved by the ethics committee of the First Affiliated Hospital of Zhejiang University. Following a full explanation of the study, written consent was obtained from each patient or his/her authorized representatives.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xia Yu and Shanshan Sun contributed equally to this work.
References
- 1.Ji Y, Ma Z, Peppelenbosch MP, Pan Q. Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob Health. 2020;8(4):e480. doi: 10.1016/S2214-109X(20)30068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody H. Influenza. Nature. 2019;573(7774):S49. doi: 10.1038/d41586-019-02750-x. [DOI] [PubMed] [Google Scholar]
- 4.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.