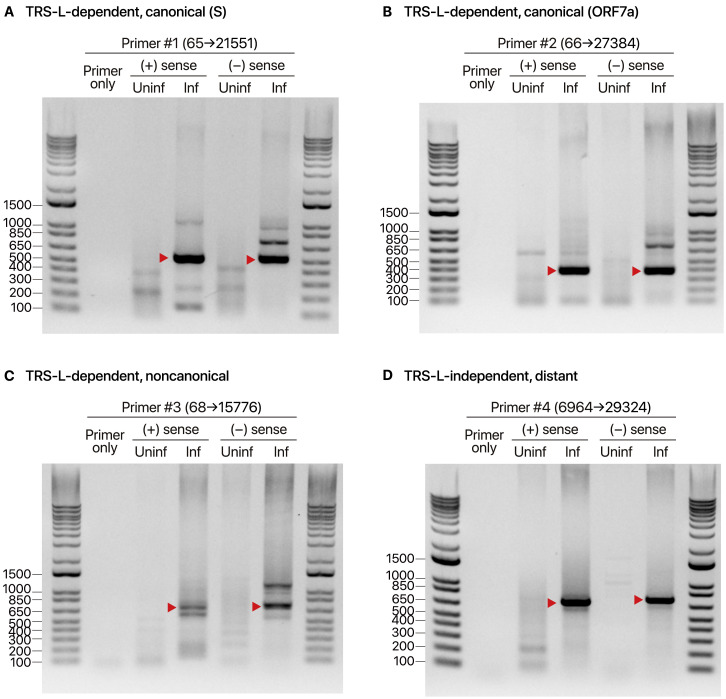
Figure S3.
Validation of Discontinuous Transcription Detected by RT-PCR, Related to Figure 3
To validate the sgRNAs found by sequencing, RT-PCR was performed to detect the sgRNAs and their negative-sense counterparts. (+) sense, cDNA from positive-strand specific reverse transcription; (−) sense, cDNA from negative-strand specific reverse transcription. ‘Primer only’ does not contain a cDNA template. cDNA from uninfected Vero cells (Uninf) were used as negative controls. Ladders are presented on the left (bp). A, RT-PCR spanning the canonical junction between TRS-L and the S ORF. B, RT-PCR spanning the canonical junction between TRS-L and the ORF7a. C, RT-PCR spanning the noncanonical junction between TRS-L and the middle of ORF1. D, RT-PCR spanning a noncanonical TRS-L-independent junction. The products were run on agarose gels. Red arrowheads denote the expected amplicons.