Abstract
Incidences of emerging/re-emerging deadly viral infections have significantly affected human health despite extraordinary progress in the area of biomedical knowledge. The best examples are the recurring outbreaks of dengue and chikungunya fever in tropical and sub-tropical regions, the recent epidemic of Zika in the Americas and the Caribbean, and the SARS, MERS, and influenza A outbreaks across the globe. The established natural reservoirs of human viruses are mainly farm animals, and, to a lesser extent, wild animals and arthropods. The intricate “host-pathogen-environment” relationship remains the key to understanding the emergence/re-emergence of pathogenic viruses. High population density, rampant constructions, poor sanitation, changing climate, and the introduction of anthropophilic vectors create selective pressure on host-pathogen reservoirs. Nevertheless, the knowledge and understanding of such zoonoses and pathogen diversity in their known non-human reservoirs are very limited. Prevention of arboviral infections using vector control methods has not been very successful. Currently, new approaches to protect against food-borne infections, such as consuming only properly cooked meats and animal products, are the most effective control measures. Though significant progress in controlling human immunodeficiency virus and hepatitis viruses has been achieved, the unpredictable nature of evolving viruses and the rare occasions of outbreaks severely hamper control and preventive modalities.
Keywords: Emerging infections, Novel viruses, Viral outbreaks, Virus evolution
Introduction
Incidences of emerging and/or re-emerging infections have significantly affected human health since ancient times [1]. The emerging pathogens are defined as novel etiological agents that have been recently introduced in a population. The “Spanish flu” responsible for tens of millions of casualties in the early twentieth century, was the most devastating natural calamity in human history [2]. The flu pandemic returned in 1957 as “Asian flu” and then in 1968 as “Hong Kong flu” that killed about three million people [2].
The most recent re-emergence of influenza on this scale was in 2009 as “swine flu” that claimed 18,500 lives [3]. On its pandemic course in 2002/2003, a novel severe acute respiratory syndrome coronavirus (SARS-CoV) infected >8,000 people, causing 774 deaths in 27 countries [4].
Actually, it was the discovery of the human immunodeficiency virus (HIV) in the early 1980s that initiated a worldwide awareness of and research interest in emerging novel viral pathogens. Over the past several decades, new outbreaks of infections have led to the discovery of a diverse array of highly pathogenic viruses mainly those belonging to the Filoviridae, Arenaviridae, Bunyaviridae, Paramyxoviridae, Coronaviridae, Flaviviridae, Togaviridae and Hepeviridae families. Some examples include BK virus (BKV), JC virus (JCV), Merkel cell polyomavirus (MCV or MCPyV), severe fever with thrombocytopenia syndrome virus (SFTSV), hantavirus (HTNV), and Sin Nombre virus (SNV) [5, 6, 7]. Following the fatal cases of Lujo virus in southern Africa in 2008 [8], another arenavirus, the Lassa virus (LASV), first reported in Nigeria in 1969, re-emerged in Guinea, Liberia, and Mali in 2009, in Ghana in 2011, and in Benin in 2014 [9].
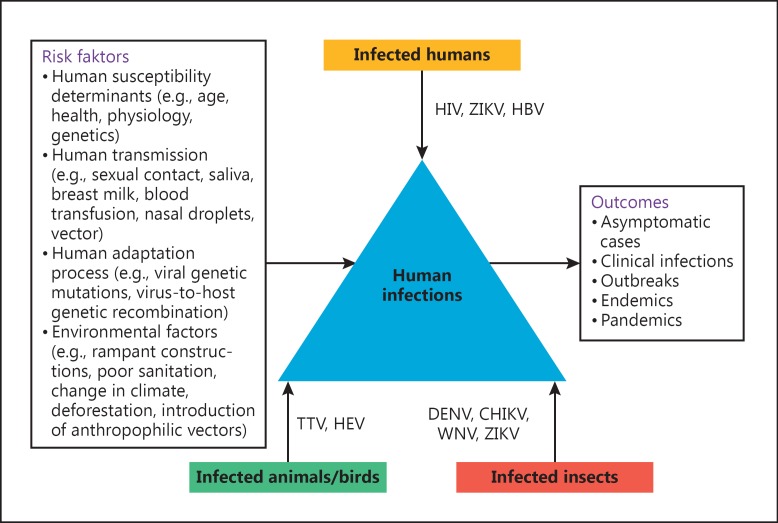
Human metapneumovirus (hMPV) was first identified in the Netherlands in 2001, and was subsequently linked to an acute lower respiratory tract infection in children, similar to respiratory syncytial virus (RSV). Recently, in 2013, a novel avian influenza A strain (H7N9) of “bird flu” in China [10] and the Middle-East respiratory syndrome (MERS)-CoV have been identified [11]. Of note, while 2015 was threatened by the re-emergence of the Ebola virus, 2015/2016 has been challenged with the resurgence of the Zika virus (ZIKV) [12]. Despite substantial advancements in the understanding of the biology of pathogens, the breakthroughs in prevention, and their effects on public health and the global economy, the emergence of novel pandemic viruses remains an enduring puzzle. This review presents an update on the knowledge of important emerging/re-emerging viral infections worldwide, discussing their possible origin, evolution, natural reservoirs, human adaptations, and risk factors (Fig. 1).
Fig. 1.
A cartoon depiction of important emerging/re-emerging viral infections and their possible origins, evolutionary drivers, natural reservoirs, and risk factors. HBV, hepatitis B virus; HEV, hepatitis E virus; HIV, human immunodeficiency virus; CHIKV, chikungunya virus; DENV, dengue virus; TTV, Torque Teno virus; WNV, West Nile virus; ZIKV, Zika virus.
Origin
Of the known viruses that infect humans, about 80% perpetuate naturally in non-human “reservoirs,” largely farm mammals and poultry and, to a lesser extent, in wild animals and arthropods [13]. It is estimated that zoonotic infectious agents constitute about 60% of the known human pathogens and up to 75% of “emerging” human pathogens [14, 15, 16]. We have, however, limited knowledge of such zoonosis and the diversity of these viruses in their known reservoirs. The data on some of the domestic mammals harboring dozens of virus species is limited and we have insufficient knowledge about the wild animals that are estimated to host thousands of virus species [17]. Examples include emerging human viruses like novel influenza strains, human (h)CoV, Hendra virus, Nipah virus, and several others, all linked to zoonosis. Recently, the deadly outbreak of MERS-CoV was linked to its zoonotic origin because of its close genetic homology to bat CoV, but not to any other known hCoV [18]. Current data shows that bats harbour the greatest diversity of CoVs, which vary from species-to-species and region-to-region [19]. The lyssaviruses of Rhabdoviridae are zoonotic human pathogens that cause fatal encephalitic disease. In addition, European bat lyssavirus type-1/-2 (EBLV-1/-2), Bokeloh bat lyssavirus (BBLV) [20], and Australian bat lyssaviruses (ABLV) have been implicated in human fatalities [21, 22, 23]. Rabies virus (RABV), the archetypal lyssavirus, is historically one of the most dreaded viruses, with zoonotic potential in dogs, cats, and ferrets, and includes other domestic and wild mammals [24].
Globally, several mammalian species, including the deer mouse, rice rat, and cotton rat are recognized as potential reservoir of HTNV [25]. In China, the HTNV and Seoul virus (SEOV) are zoonotically linked to the striped field mouse and the Norway rat, respectively. Also in China, the Fugong virus (FUGV), a novel HTNV has recently been identified in small oriental voles [26]. HTNVs have also been detected in shrews and bats, but their link to human illness remains to be established.
Moreover, in addition to humans and pigs, there is a growing chain of mammalian hosts for the hepatitis E virus (HEV) that includes deer, boar, mongoose, rabbit, rat, goat, camel, bat, ferret, moose, etc. [27, 28]. Likewise, the highly prevalent Torque Teno virus (TTV), of the newly established virus family, Anelloviridae, also infects pigs, cows, sheep, cats, dogs, and chickens [29].
Arboviruses like dengue virus (DENV), chikungunya virus (CHIKV), ZIKV, and West Nile virus (WNV) are the arthropod-borne viruses that have re-emerged in many tropical and subtropical regions in the last 2 decades. Notably, ZIKV was known as a neglected tropical disease confined to Africa and Asia until an outbreak was reported in 2007 on Yap Island and in 2013/2014 on islands in the Pacific, thus expanding its geographical territory [30, 31]. WNV remains the most important mosquito-borne encephalitis pathogen in North America, involving Culex sp. and the American Robin in its transmission cycle. Since its emergence in the West in 1999, it has undergone adaptive genetic changes as it spreads throughout North America [32]. Furthermore, Crimean-Congo hemorrhagic fever virus (CCHFV) is considered to be one of the major emerging diseases spreading to and within the European nations, following the expanding distribution of anthropophilic ticks [33]. Every year, >1,000 cases of CCHFV, due to human-to-human transmission, are reported in southeastern Europe, including Turkey. SFTSV, a previously unidentified tick-borne bunyavirus, has recently emerged in China with fatality rates as high as 30% [34].
Evolution and Adaptation to the Human Host
While DNA viruses are believed to have been evolving and diversifying for millions of years [35], most RNA viruses are likely to have a much more recent evolution and “human-adaptation” for only thousands of years [36, 37]. Owing to their error-prone polymerase/reverse-transcriptase (approx. 10-4/site/replication cycle) that seems to operate at optimal fidelity, RNA viruses exist as more genetically diversified populations than DNA viruses. Nevertheless, of the hitherto recognized 158 species of human RNA viruses consisting of 47 genera and 17 families, only a minority has adapted to humans [38]. In contrast, of the known 91 DNA virus species with 22 genera and 8 families, nearly 87% are adapted to human hosts [38]. In the human-adaptation process, the viral genetic mutations, re-assortment or virus-host genetic recombination might lead to the establishment of stable virus lineages in human populations. It is, therefore, quite possible that such human-adapted viruses could circulate asymptomatically and remain undetected until their novel clinical manifestations are noticed. To understand this further, a recently isolated HEV genotype 3 from a chronic hepatitis E patient containing a recombinant virus-host RNA genome was shown to infect cultured human, pig, and deer hepatocytes [39].
It is assumed that there is a pool of human virus species still to be discovered. The composition of such a viral pool is dynamic, changing over time, i.e., while some virus species tend to become extinct, others continue to evolve in their natural hosts. More commonly, new species arise as a result of jumps from one host to another, thus crossing the species barrier [36]. Humans are, therefore, no more than “incidental” or “spillover” hosts for pathogens. However, only a minority of such viruses are capable of persisting in certain human populations (endemics) or spreading across populations (epidemics) in the absence of a reservoir.
Differential host factors like age, health, physiology, nutritional status, exposure history, simultaneous infection with >1 pathogen, immunocompetence, and genetics are determinants to human susceptibility to an infection. The field of phylodynamics, combining a modeling framework for host, epidemiological, and molecular data, especially for RNA viruses, shows particular promise for understanding the patterns of viral evolution during epidemics [40, 41]. Moreover, our underappreciated aspect of growing human populations, global changes in land usage, and the introduction of anthropophilic vectors create selective pressure on hosts and reservoirs. For example, both WNV and CHIKV evolved rapidly after being introduced to new locations and encountering new vectors. When Aedes albopictus rather than A. aegypti became the main vector of CHIKV in the Indian subcontinent after the 2004-2009 epidemic, the same viral strain spread rapidly, and the subsequent mutants seemed to circulate and persist more efficiently [42]. A unique molecular signature of the CHIKV epidemic was a single amino acid substitution (A226V) in the envelope protein (E1). This mutation was identified in 90% of the strains reported during the later phase, and was associated with a high epidemic potential for CHIKV [43]. ZIKV, initially known to be transmitted by mosquitoes, was recently reported as being transmitted, albeit more rarely, via sexual contact, saliva, breast milk, blood transfusions, and from mother to child [12, 44].
Evolutionary Drivers
The key to understanding the emergence/re-emergence of novel viruses is to know the intricate “host-pathogen-environment” relationship in the evolution of pathogens. While the emergence of infectious diseases in naive regions is caused primarily by the movement of pathogens via trade and travel, local emergence is driven by a combination of environmental and social change. Notably, virus transmission rates are often higher in dense than in sparse populations, and the spread is often greatly enhanced by air travel or migration. Pathogens introduced into novel regions often cause explosive epidemics followed by a declining incidence whereas those that emerge locally due to land usage or social changes usually show consistent increases. A recent example is the emergence of ZIKV in Brazil in 2015 [12, 44]. Phylogenetic studies suggested that ZIKV from the Pacific islands outbreak in 2013/2014 was probably introduced into Brazil during the FIFA World Cup or the 2014 FIA World Endurance Championship auto racing series. The dispersal of the virus in Brazil resulted in an explosive epidemic of Zika fever, and the infection spread to other countries due to frequent travel.
While most human infections are known to have zoonotic origins, it is certain that alterations in the environment, due to industrialization and urbanization, are an important but completely neglected factor. Though the origins of most of the human viruses are not known so far, the great majority can be categorized as “crowd diseases” that require a relatively high host-density to persist [35]. Notably, the recent outbreaks of H1N1, hCoV, Hendra virus, Nipah virus, and MERS-CoV suggest that the Asia-Pacific region is the global hot-spot for the emergence of novel RNA viruses. In this case, an order-of-magnitude estimation of 1 such event per 100 years is broadly consistent with human demographic history [37].
Notably, RNA viruses are known to incorporate drastic mutations in their genome, an example being the large duplication events in the G protein gene of RSV. Two such remarkable events were the 60-bp duplication in group B RSV in Argentina in 1999 and the 72-bp duplication in group A RSV in Canada in 2011 [45, 46]. These new genotypes of RSV with their duplications, known as BA and ON1, respectively, spread to different geographical regions across the globe due to immunologically naive travellers [47, 48, 49, 50]. The mathematics of these spreading events is well known today, and a sophisticated array of computational and mathematical models can be used to accurately back-predict such events. An example of this is the first case-clusters of the SARS outbreak and the subsequent global spread, including the country-by-country distribution of human cases [51, 52]. Recent investigations have evaluated the transmission dynamics of ZIKV infection using mathematical models [53].
We may not ignore that wild animals constitute an important but poorly understood reservoir for known and undiscovered human pathogens, including viruses. Furthermore, the relative importance of an animal species as a source of human infection is a function of the prevalence of zoonotic agents in that species and the probability of close contact (direct or indirect) with susceptible humans. Clearly, these factors vary geographically, and changes in patterns of human and animal disease will continue to result from socio-economic and ecological changes at the human-to-animal interface.
Potential Risks and Control Measures
If risk is a function of frequency of contact and a probability of successfully adapting to human hosts, viruses acquired from non-human primates might already be better adapted to successful transmission than those from other mammals, like bovine, porcine, feline, and rodent mammals. For example, SARS-CoV has been detected in masked-palm or gem-faced civet cats which are sold at Chinese wildlife markets [54, 55]. Like HEV, many enteric viruses (e.g., JCV) are found worldwide in high concentrations in urban sewage, and lead to water contamination in countries with poor resources [56].
Recurring epidemics caused by emerging viruses have necessitated the formulation of effective control measures, but the zoonotic transmission of many of these viruses has contributed significantly towards the challenges associated with their prevention and control. For instance, the prevention of arthropod-borne infections (e.g., DENV, CHIKV, and ZIKV) using vector control methods has only been partially successful [57]. Preventing mosquitos breeding can be mediated by avoiding the accumulation of rain water in endemic regions. Other preventive measures like the use of repellent creams and clothes that cover the body should be employed to avoid mosquito bites. Insectidal sprays should be utilized in endemic regions to control the mosquito population.
Additionally, prevention of the transmission of other zoonotic viruses should also be addressed in a systematic manner to combat the infection of humans with these pathogens. It is now well known that many emerging human viruses can be transmitted via contact with infected animals and the consumption of animal products, including freshwater and seafood products. The new approach to food safety and to protecting humans against food-borne health risks is the most effective measure to control the human illnesses associated with animal diseases. However, the vast majority of human infections are not reported to clinicians, and so such etiological agents remain unidentified and continue to contaminate the population. As discussed, poultry are high-risk birds for emerging novel influenza A strains. Pork is another potential source of chronic HEV. In a public health care initiative, the French Health Authority has recommended to cook pig-liver sausages prior to consumption [58]. Precautions and proper care should therefore be taken when selecting, purchasing, and hunting high-risk animals and when cooking meat. The current biosecurity measures appear to have been generally more successful for constraining bacterial diseases, but less effective for viral diseases. Surveillance of the emerging human viruses must include livestock, wild animal, and potential arthropod vectors as well as their environments at an international level of coordination. Furthermore, individuals like livestock herders, zookeepers, hunters, rangers, and veterinarians working with reservoir or high-risk animals must take hygienic measures.
Notably, many viruses that affect the respiratory tract, like influenza, RSV, MERS-CoV, SARS-CoV, and hMPV are transmitted by respiratory secretions and aerosol droplets. Effective measures for preventing their transmission in a community and their nosocomial spread are the adoption of good practices like frequent hand-washing and avoiding direct contact with patients [59]. In addition, generating awareness in the general public about various aspects of these diseases will also assist in the better management and control of these emerging viral infections.
Current Challenges
The overall balance between exposure and evolution as driving forces of human virus diversity is difficult to assess accurately, particularly for viruses that are not known to have adapted to humans and that exist primarily in animal reservoirs. Despite landmark advances in understanding the nature and biology of many pathogenic viruses, there is limited knowledge on emerging novel viruses, their potential reservoirs, and their modes of transmission. For example, although the person-to-person transmission of BKV is known, no zoonotic or other origin has yet been established. Likewise, though MCV is found in respiratory secretions, healthy skin sheddings and gastrointestinal tissues, its precise mode of transmission still remains undetermined. The perpetual nature of the emergence and transmission of infectious diseases therefore poses an ongoing challenge which is volatile and ever-changing. However, despite incidences of infection and pathogenicity, for many viruses like CCHFV and ZIKV, there is no vaccine prophylaxis or therapeutic intervention available at present. The recurring post-monsoon outbreaks of dengue and chikungunya fever and their control in the Indian subcontinent remain challenging. The complexity of the co-circulation of DENV, CHIKV, and ZIKV in many regions makes differential diagnosis difficult due to overlapping clinical manifestations and the partial cross-reactivity of antibodies [60, 61, 62, 63]. These challenges include a requirement for constant surveillance, prompt and efficient diagnosis, and development of new therapies. There is a further need for devoted research, not only to develop counter-measures but also to understand the basic biology of new viral pathogens and human susceptibilities. While advances in biomedical technologies have enabled us to prospectively identify pathogenic inter-species viruses and assess their transmissibility, the environmental release of several potential “laboratory-adapted” viruses is considered to be a big threat.
Conclusions and Future Prospects
The frequent emergence or re-emergence of deadly viral infections has significantly affected human health despite extraordinary progress in biomedical knowledge. High population density, rampant constructions, poor sanitation, changing climate, and the introduction of anthropophilic vectors create selective pressure on hosts and pathogen reservoirs. Although we know that a substantial fraction of well-identified mammalian viruses is responsible for human etiology, there are large numbers of evolving viruses waiting to infect and adapt. The unpredictable nature of novel infections, the rare occasions of outbreaks, the small numbers of confirmed cases, the asymptomatic occurrence, and occurrence in remote areas severely hamper the assessment of control and preventive modalities. Efficient vaccines are already available against important human viruses, like HBV and human papillomavirus. Even though drug resistance in HBV and HIV has accelerated alarmingly, new generations of broad-spectrum anti-viral agents as combination regimens hold promise. Moreover, specific geographical regions or interfaces between humans, livestock, wildlife, and the environment should be targets for intense surveillance. In this way, further extensive research will surely enhance our understanding and capacity for predicting new pandemics and preparing control measures in advance.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgement
M.K.P. gratefully acknowledges the intellectual support and guidance of his mentor Dr. Shahid Jameel (CEO, Wellcome Trust-UK/DBT-India Alliance).
References
- 1.Krause RM. The origin of plagues: old and new. Science. 1992;257:1073–1078. doi: 10.1126/science.257.5073.1073. [DOI] [PubMed] [Google Scholar]
- 2.Wever PC, van Bergen L. Death from 1918 pandemic influenza during the First World War: a perspective from personal and anecdotal evidence. Influenza Other Respir Viruses. 2014;8:538–546. doi: 10.1111/irv.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawood FS, Iuliano AD, Reed C, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 4.WHO: Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003 http://www.who.int/csr/sars/country/table2004_04_21/en/ [Google Scholar]
- 5.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 6.Padgett BL, Walker DL, ZuRhein GM, et al. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 7.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paweska T, Sewlall NH, Ksiazek TG, et al. Nosocomial outbreak of novel arenavirus infection, southern Africa. Emerg Infect Dis. 2009;15:1598–1602. doi: 10.3201/eid1510.090211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO: Lassa fever fact sheet 2016 http://www.who.int/mediacentre/factsheets/fs179/en/.
- 10.Lu S, Xi X, Zheng Y, et al. Analysis of the clinical characteristics and treatment of two patients with avian influenza virus (H7N9) Biosci Trends. 2013;7:109–112. [PubMed] [Google Scholar]
- 11.Khan G. A novel coronavirus capable of lethal human infections: an emerging picture. Virol J. 2013;10:66–65. doi: 10.1186/1743-422X-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musso D, Gubler DJ. Zika Virus. Clin Microbiol Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil Trans R Soc Lond B. 2011;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Phil Trans R Soc Lond B. 2013;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and re-emerging pathogens. Emerg Infect Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilpatrick AM, Randolph SE. Drivers, dynamics, and control of emerging vector-borne zoonotic diseases. Lancet. 2012;380:1946–1955. doi: 10.1016/S0140-6736(12)61151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cleaveland S, Haydon DT, Taylor L. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr Top Microbiol Immunol. 2007;315:85–111. doi: 10.1007/978-3-540-70962-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corman VM, Eckerle I, Bleicker T, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:pii–20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 19.Anderson LJ, Tong S. Update on SARS research and other possibly zoonotic coronaviruses. Int J Antimicrob Agents. 2010;36:S21–S25. doi: 10.1016/j.ijantimicag.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McElhinney LM, Marston DA, Leech S, et al. Molecular epidemiology of bat lyssaviruses in europe. Zoo Pub Health. 2013;60:35–45. doi: 10.1111/zph.12003. [DOI] [PubMed] [Google Scholar]
- 21.McCall BJ, Epstein JH, Neill AS, et al. Potential exposure to Australian bat lyssavirus, Queensland, 1996-1999. Emerg Infect Dis. 2000;6:259–264. doi: 10.3201/eid0603.000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allworth A, Murray K, Morgan J. A human case of encephalitis due to a Lyssavirus recently identified in fruit bats. Commun Dis Intell. 1996;20:504–510. [Google Scholar]
- 23.Hanna JN, Carney IK, Smith GA, et al. Australian bat lyssavirus infection: a second human case, with a long incubation period. Med J Aust. 2000;172:597–599. doi: 10.5694/j.1326-5377.2000.tb124126.x. [DOI] [PubMed] [Google Scholar]
- 24.Fooks A. The challenge of new and emerging lyssaviruses. Expert Rev Vaccines. 2004;3:333–336. doi: 10.1586/14760584.3.4.333. [DOI] [PubMed] [Google Scholar]
- 25.CDC: Rodents in the United States that carry hantavirus http://www.cdc.gov/hantavirus/rodents/index.html.
- 26.Ge X, Yang W, Pan H, et al. Fugong virus, a novel hantavirus harbored by the small oriental vole (Eothenomys eleusis) in China. Virol J. 2016;13:27–32. doi: 10.1186/s12985-016-0483-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavio N, Meng XJ, Doceul V. Zoonotic origin of hepatitis E. Curr Opin Virol. 2015;10:34–41. doi: 10.1016/j.coviro.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Widén F. Hepatitis E as a zoonosis. Adv Exp Med Biol. 2016;948:61–71. doi: 10.1007/978-94-024-0942-0_4. [DOI] [PubMed] [Google Scholar]
- 29.Spandole S, Cimponeriu D, Berca LM, Mihăescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol. 2015;160:893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 30.Duffy M R, Chen T-H, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. New Eng J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 31.PAHO: Regional Zika epidemiological update (Americas) 2016 http://www.paho.org/hq/index.php?option=com_content&id=11599&Itemid=41691. [Google Scholar]
- 32.Pesko KN, Ebel GD. West Nile virus population genetics and evolution. Infect Genet Evol. 2012;12:181–190. doi: 10.1016/j.meegid.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertens M, Schmidt K, Ozkul A, Groschup MH. The impact of Crimean-Congo hemorrhagic fever virus on public health. Antiviral Res. 2013;98:248–260. doi: 10.1016/j.antiviral.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Yu XJ, Liang MF, Zhang SY, et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmonds P. Reconstructing the origins of human hepatitis viruses. Phil Trans R Soc Lond B. 2011;356:1013–1026. doi: 10.1098/rstb.2001.0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitchen A, Shackelton LA, Holmes EC. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc Natl Acad Sci USA. 2011;108:238–243. doi: 10.1073/pnas.1011090108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolhouse MEJ, Adair K. The diversity of human viruses. Future Virol. 2013;8:159–171. doi: 10.2217/fvl.12.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla P, Nguyen HT, Torian U, et al. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci USA. 2011;108:2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grenfell BT, Pybus OG, Gog JR, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 41.Holmes EC. The Evolution and Emergence of RNA Viruses. Oxford: Oxford University Press; 2009. [Google Scholar]
- 42.Tsetsarkin KA, Weaver SC. Sequential adaptive mutations enhance efficient vector switching by chikungunya virus and its epidemic emergence. PLoS Pathog. 2011;7:e1002412. doi: 10.1371/journal.ppat.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abushouk AI, Negida A, Ahmed H. An updated review of Zika virus. J Clin Virol. 2016;84:53–58. doi: 10.1016/j.jcv.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Trento A, Galiano M, Videla C, Carballal G, Garcia-Barreno B, et al. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol. 2003;84:3115–3120. doi: 10.1099/vir.0.19357-0. [DOI] [PubMed] [Google Scholar]
- 46.Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, et al. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One. 2012;7:e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed A, Haider SH, Parveen S, et al. Co-circulation of 72-bp duplication group A and 60-bp duplication group B respiratory syncytial virus (RSV) strains in Riyadh, Saudi Arabia during 2014. PLoS One. 2016;11:e0166145. doi: 10.1371/journal.pone.0166145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duvvuri VR, Granados A, Rosenfeld P, Bahl J, Eshaghi A, et al. Genetic diversity and evolutionary insights of respiratory syncytial virus A ON1 genotype: global and local transmission dynamics. Sci Rep. 2015;5:14268. doi: 10.1038/srep14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trento A, Casas I, Calderon A, Garcia-Garcia ML, Calvo C, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 2010;84:7500–7512. doi: 10.1128/JVI.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parveen S, Sullender WM, Fowler K, Lefkowitz EJ, Kapoor SK, Broor S. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J Clin Microbiol. 2006;44:3055–3064. doi: 10.1128/JCM.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson RM, Fraser C, Ghani AC, et al. Epidemiology, transmission dynamics and control of SARS: the 2002-2003 epidemic. Phil Trans R Soc Lond B. 2004;359:1091–1105. doi: 10.1098/rstb.2004.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hufnagel L, Brockmann D, Geisel T. Forecast and control of epidemics in a globalized world. Proc Natl Acad Sci USA. 2004;101:15124–15129. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kucharski AJ, Funk S, Eggo RM, et al. Transmission dynamics of Zika virus in island populations: a modeling analysis of the 2013-14 French Polynesia outbreak. PLoS Negl Trop Dis. 2016;10:e0004726. doi: 10.1371/journal.pntd.0004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li WD, Shi ZL, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 55.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 56.Bofill-Mas S, Formiga-Cruz M, Clemente-Casares P, et al. Potential transmission of human polyomaviruses through the gastrointestinal tract after exposure to virions or viral DNA. J Virol. 2001;75:10290–10299. doi: 10.1128/JVI.75.21.10290-10299.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deeba F, Islam A, Kazim SN, Naqvi IH, Broor S, Ahmed A, Parveen S. Chikungunya virus: recent advances in epidemiology, host pathogen interaction and vaccine strategies. Pathog Dis. 2016;74:ftv119. doi: 10.1093/femspd/ftv119. [DOI] [PubMed] [Google Scholar]
- 58.Parvez MK. Chronic hepatitis E infection: risks and controls. Intervirol. 2013;56:213–216. doi: 10.1159/000349888. [DOI] [PubMed] [Google Scholar]
- 59.French CE, McKenzie BC, Coope C, et al. Risk of nosocomial respiratory syncytial virus infection and effectiveness of control measures to prevent transmission events: a systematic review. Influenza Other Respir Viruses. 2016;10:268–290. doi: 10.1111/irv.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afreen N, Naqvi IH, Baroor S, Ahmed A, Parveen S. Phylogenetic and molecular clock analysis of dengue serotype 1 and 3 from New Delhi, India. PLoS One. 2015;10:e0141628. doi: 10.1371/journal.pone.0141628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tazeen A, Afreen N, Abdullah M, et al. Occurrence of co-infection with dengue viruses during 2014 in New Delhi, India. Epidemiol Infect. 2016;13:1–11. doi: 10.1017/S0950268816001990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lahariya C, Pradhan S. Emergence of chikungunya virus in Indian subcontinent after 32 years: a review. J Vector borne Dis. 1996;43:151–157. [PubMed] [Google Scholar]
- 63.Afreen N, Deeba F, Khan WH, et al. Molecular characterization of dengue and chikungunya virus strains circulating in New Delhi, India. Microbiol Immunol. 2014;58:688–696. doi: 10.1111/1348-0421.12209. [DOI] [PubMed] [Google Scholar]