Abstract
Turkey coronavirus (TCoV) polyprotein was predicted to be cleaved into 15 non-structural proteins (nsp2 to nsp16), but none of these nsps have been characterized. TCoV nsp15 consists of 338 residues and shares 40% sequence similarity to U-specific Nidovirales endoribonuclease (NendoU) of severe acute respiratory syndrome coronavirus.
Objective
The purpose of the present study was to characterize TCoV nsp15.
Methods
The TCoV nsp15 gene was cloned into pTriEX1 and expressed as a C-terminal His-tagged recombinant protein in BL21 (DE3). The recombinant nsp15 was purified by Ni-NTA resin. Synthetic RNA substrates were used to determine the substrate specificity of the TCoV nsp15. RNA zymography was used to determine the active form of the nsp15.
Results
The TCoV nsp15 did not cleave DNA but degraded total cellular RNA. The TCoV nsp15 cleaved single-stranded (ss) RNA at the uridylate site. The TCoV nsp15 cleaved hairpin RNA, pRNA, and double-stranded RNA (dsRNA) of infectious bursal disease virus very slowly, implying that dsRNA is not a good substrate for the TCoV nsp15. No divalent metal ion was required for in vitro enzymatic activity of the TCoV nsp15. The active form of the TCoV nsp15 was a homohexamer and disulfide bond was essential for the enzymatic activity.
Conclusion
The TCoV nsp15 is a NendoU but has some characteristics different from other NendoU.
Key Words: Turkey coronavirus, nsp15, U-specific Nidovirales endoribonuclease, Endoribonuclease
Introduction
Coronavirus consists of a group of viruses infecting both humans and animals and causing mostly respiratory disease [1]. The family Coronaviridae contains more than 10 coronaviruses which are classified into three groups based on their antigenic and phylogenetic relationships. Together with the families Arteriviridae and Roniviridae, Coronaviridae is classified as a member in the order Nidovirales [2]. The coronavirus is enveloped and corona-like spikes or peplomers can be seen around the envelope when viewed under electron microscopy. The core inside the envelope contains a single-stranded (ss), positive-sense genome RNA of 27–33 kb wrapped by nucleocapsid protein. The coronavirus RNA genome is the largest RNA genome among all known non-segmented RNA viruses and encodes more than 10 AUG-containing open reading frames (ORF), each of which is expressed as an independent protein [3]. About two thirds of the genome at the 5′ end encodes two ORFs, 1a and 1b, which are translated to produce polyproteins pp1a and pp1ab, the latter being produced through a −1 frameshift translational mechanism [3, 4, 5]. Structural genes are located at the 3′ end of the genome. One unique feature of coronaviruses is that they produce a set of 5′ and 3′ co-terminal subgenomic mRNAs (sgRNA) for structure and accessory gene production. The mechanism for sgRNA production is currently under debate [6, 7].
Coronavirus polyprotein pp1ab is a large polypeptide. The C-terminal part of the pp1ab shares relatively high conservation among coronaviruses. Polyprotein pp1ab is processed into more than 15–16 non-structural proteins (nsps) by polyprotein-encoded 3-cystine-like proteinase (3CLpro) and papain-like proteinase (PLP). These nsps form replication complexes where coronavirus genome replication and transcription take place. Bioinformatics analysis predicted the existence of another six enzymes, including RNA-dependent RNA polymerase (RdRp), NTPase/helicase, exoribonuclease (ExoN), endoribonuclease (endoRNAse), 2′-O-methyltransferase (2′-O-MT) and ADP-ribose-1′-phosphatase diesterase [8]. Among these enzymes, endoribonuclease has gained much attention since the discovery that the endoribonuclease activity of severe acute respiratory syndrome (SARS) nsp15 is similar to that of Xenopus XendoU [9], which requires manganese as a cofactor to cleave uridylate-containing ssRNA [10, 11]. Due to the sequence conservation of SARS nsp15 within the order Nidovirales, U-specific Nidovirales endoribonuclease (NendoU) was engineered to refer to Nidovirales endoU to indicate its relation with XendoU [11]. The activity of recombinant NendoU has been identified for human coronavirus 229E (HCoV-229E) nsp15 [11], mouse hepatitis virus (MHV) A59 nsp15 [12], and equine arteritis virus (EAV) nsp11 [13]. Unlike XendoU, which is a monomer that cleaves ssRNA, NendoU functions as a hexamer that cleaves both ss- and double-stranded (ds)-RNA [12, 14, 15, 16]. Reverse genetics with point mutation revealed that NendoU might be involved in genome replication and sgRNA transcription, but its exact role requires further investigation [11, 13].
Turkey poult enteritis is caused by infection with turkey coronavirus (TCoV) and is characterized by acute atrophic enteritis and reduced weight gain in turkeys [17]. Outbreaks of TCoV infection have resulted in significant economic loss in the US turkey industry [18, 19]. TCoV, along with infectious bronchitis virus (IBV) in chickens, is a group 3 coronavirus within the family Coronaviridae. The whole genome of TCoV is 27.7 kb and encodes 13 ORFs [20]. Polyprotein mapping predicted the presence of 14 nsps for pp1ab in which nsp15 shared 40% sequence similarity with nsp15 of SARS-CoV and other coronaviruses and was predicted to be NendoU [20]. The objective of the present study was to further characterize the enzymatic activity and substrate specificity of the TCoV nsp15.
Materials and Methods
Construction of Expression Plasmid
Primers nsp15F (CTAGccatggcgTCTATTGATAATATTGCT) and nsp15R (GCTAggtaccTTGAAGCTGTGGATAACA) were used to amplify TCoV nsp15 from cDNA of the TCoV 540 strain. The underlined sequences are restriction sites used for cloning the gene into the expression vector pTriEX1.1 (Novagen, Gibbstown, N.J., USA) to construct plasmid pTrinsp15 for the production of C-terminal His-tagged fusion protein. The PCR reaction consisted of 1× PCR buffer (Promega, Madison, Wisc., USA), 200 μμ each of dNTPs, 200 nm of nsp15F and nsp15R, 2 μl of cDNA, and 2.5 μ of Taq polymerase in a final volume of 50 μl. PCR conditions were as follows: initial denaturation at 94° for 3 min followed by 30 cycles of denaturation at 93° for 10 s, annealing at 55° for 30 s, and extension at 72° for 1 min. A final extension at 72° for 10 min was included before the completion of the reaction. The PCR product was analyzed by agarose gel electrophoresis and purified by Zymo DNA Clean & Concentrator-5 (Zymo Research, Orange, Calif., USA). Purified PCR product was digested with NcoI and KpnI overnight and purified again. 2 μl of purified digestion products were cloned into pTriEX1.1, which was treated in the same way as for the nsp15 PCR product. The ligation reaction was carried out at 16° overnight, and 2 μl of the reaction was transformed into TOP10 chemically competent cells according to the vendor's procedure (Invitrogen, Carlsbad, Calif., USA). Positive colonies were screened by PCR and enzyme digestion. Plasmid pTrinsp15 was isolated with QIAprep spin miniprep kit (Qiagen, Valencia, Calif., USA) and submitted for DNA sequencing at the Genomic Core Facility, Purdue University. The plasmid was then transformed into BL21 (DE3) chemically competent cells for nsp15 expression. The predicted fusion nsp15 contained 377 amino acids (aa) including 338 aa from nsp15. The predicted size of the recombinant nsp15 is 42.56 kDa.
Protein Expression and Purification
A single colony of BL21 (DE3) containing plasmid pTrinsp15 was cultured overnight in 3 ml of 2× YT medium containing 100 μg/ml ampicillin at 37°, 250 rpm. On the following day, the whole culture was inoculated into 100 ml of 2× YT medium in the same conditions as the overnight culture. After growth for 3 h (OD600 between 0.5 and 0.8), IPTG was added to a final concentration of 0.5 mM. Induction was continued for another 3 h. Cells were then harvested and nsp15 was purified using a Ni-NTA mini column according to manufacturer's procedure (Novagen) with some modifications. Cells were lysed in cold lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) containing 1 mM PMSF (Roche, Indianapolis, Ind., USA) and 0.5% Triton X-100. Cell lysate was cleared for 30 min at 15,000 rpm (JA25.5, Beckman) at 4°. Supernatant was transferred to a new tube and imidazole was added to a final concentration of 15 mM. The supernatant was passed through a Ni-NTA mini column pre-equilibrated with binding buffer (lysis buffer with 15 mM imidazole) and then washed with binding buffer containing 15 and 50 mM imidazole, respectively. nsp15 was eluted in binding buffer containing 400 mM imidazole and collected at 0.5 ml per tube. The concentration of collected protein was measured and the peak tubes were pooled. The pooled samples were concentrated and the buffer was changed to lysis buffer containing 50% glycerol.
The purified nsp15 was aliquoted into 100 μl per tube and stored at −20° before use. Protein concentration was measured using Bio-Rad's reagent with BSA as the standard. SDS-PAGE and Western blotting with mouse anti-His mAb were used to confirm the purified nsp15.
Enzymatic Activity Assay
The standard enzymatic assay was as follow: 1× assay buffer (50 mM Tris-HCl, pH 7.5;0.15 M NaCl; 0.1 M KCl; 5 mM Mn2+) containing 1 μmol RNA substrate and 100 ng of purified nsp15. The reaction was incubated at 37° for 30 min or as indicated. After incubation, the reaction was stopped by adding 2× RNA loading buffer and heated for 3 min at 75°. The samples were chilled on ice immediately and separated on 1% agarose gel (for RNA substrates >100 bp) or 24% PAGE gel containing 6 M urea (for RNA oligo) to separate the RNA substrate from its products. To determine the effect of divalent metal ions on nsp15 activity, buffers were also prepared in the absence of Mn2+. Different divalent ions were added to the buffer to give a final concentration of 5 mM. In the EDTA chelating assay, nsp15 was incubated in the 1× assay buffer (without Mn2+) containing different concentrations of EDTA for 10 min on ice, then RNA substrate was added, and the reaction was incubated for 30 min at 37°. About 1 μg of infectious bursal disease virus (IBDV) dsRNA was incubated with 100 ng of the nsp15 for different times and separated on 1.2% agarose gel without denaturation. The undigested dsRNA was quantified by densitometry.
RNA Zymography
10% SDS-PAGE gel was cast to contain 0.1 mg/ml yeast RNA. nsp15 (0.1 μg) was dissolved in 1× SDS loading buffer with or without β-mercaptoethanol (β-ME) and denatured at 95° for 5 min. SDS-PAGE was carried out at 160 V for 45 min. After electrophoresis, gels were incubated in a clean plastic tray and washed quickly with distilled water three times. The gel was then incubated for 30 min at room temperature with 40 ml of 25% isopropanol in 20 mM Tris-HCl, pH 7.5. This incubation was repeated once. The gel was then rinsed in distilled water three times and incubated in 40 ml reaction buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) overnight at room temperature. The gel was finally incubated in water containing 0.1 μg/ml ethidium bromide for 10 min to view the digested RNA.
Substrates for nsp15
Total cellular RNA was isolated from baby hamster kidney (BHK) cells with RNApure agent (GenHunter, Nashville, Tenn., USA) and was used at 5 μg per reaction. Primer N102F (AGTAGAGGCGGAAGAAAACCAGTC) served as ssDNA substrate. dsDNA was a PCR product of TCoV cDNA amplified with N102F and N102R (ACGCCCATCCTTAATACCTTCCTC) primers. Synthetic RNA substrates were from Dharmacon or made in vitro using T7 RNA polymerase (Promega). The sequences of the RNA substrates are listed in table 1. Hairpin RNA and pRNA were kindly provided by Dr. Peixuan Guo (Department of Comparative Pathobiology, Purdue University).
Table 1.
Oligos used for RNA substrate synthesis
| Oligo | Sequence (5′ to 3′) of DNA | Sequence of RNA (5′ to 3′) |
|---|---|---|
| PolyU50 | A50 C TAA TAG TGA GTC GTA TTA | 5′ AGU50 |
| PolyA50 | T50 C TAA TAG TGA GTC GTA TTA | 5′ AGA50 |
| TJ1 | (CCAAGG)4 | |
| TJ2 | CAGCAGUUCAGCAG |
Results
TCoV nsp15 Protein Expression and Purification
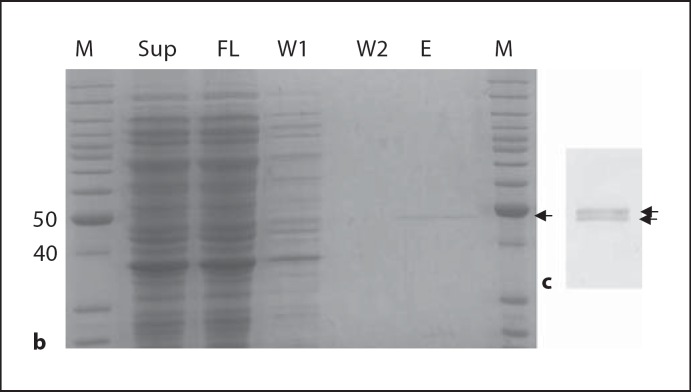
TCoV nsp15 was amplified from cDNA of the TCoV 540 isolate and cloned into the pTriEX vector. The deduced amino acid sequence of the TCoV nsp15 is shown in figure 1a in alignment with nsp15s from other coronaviruses. From the alignment, it was found that important residues associated with nsp15 activity are conserved among the nsp15 polypeptides. To express the recombinant TCoV nsp15, plasmid pTrinsp15 was transformed into BL21 (DE3) cells. A typical expression pattern of TCoV nsp15 in BL21 (DE3) is shown in figure 1b. After 3 h of induction with IPTG, there was no obvious nsp15 expression (fig. 1b). After purification through Ni-NTA resin, a single band around 43 kDa was seen on SDS-PAGE. Western blotting with mouse anti-His mAb revealed that this band was His-tagged and was thus the expected fusion nsp15. We used this purified nsp15 for all other experiments.
Fig. 1. a.
Amino acid sequence alignment of TCoV nsp15 with selected nsp15 sequences from other coronaviruses. * Denotes key residues associated with NendoU activity. b, c Expression of TCoV nsp15: b SDS-PAGE analysis of nsp15 purification from BL21 (DE3) cells. M = Molecular weight marker (Invitrogen); Sup = cleared cell lysate; FL = flow-through after passing through Ni-NTA column; W1 and W2, washes; E, elutes; c Western blot of purified nsp15 with mouse anti-His mAb and detected with goat anti-mouse IgG-HRP. Arrows indicate the nsp15.
RNase Activity of TCoV nsp15
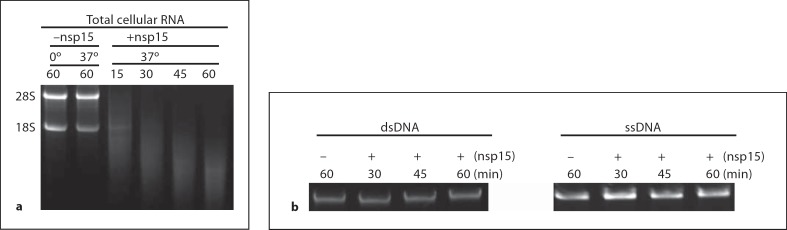
figure 2a shows that total RNA from BHK cells was rapidly degraded by nsp15. The degraded RNA formed smears, and as incubation time progressed, the degraded RNA became smaller and smaller, indicating that the total RNA was randomly degraded by nsp15. However, nsp15 did not degrade either ssDNA or dsDNA (fig. 2b), supporting characterization of nsp15 as a ribonuclease.
Fig. 2.
TCoV nsp15 activity toward total RNA and DNA. a Cleavage of cellular total RNA with TCoV nsp15 at 37° for increasing amounts of time. b Cleavage of dsDNA (left) and ssDNA (right) by TCoV nsp15 at 37° with different periods of incubation.
Substrate Specificity of TCoV nsp15
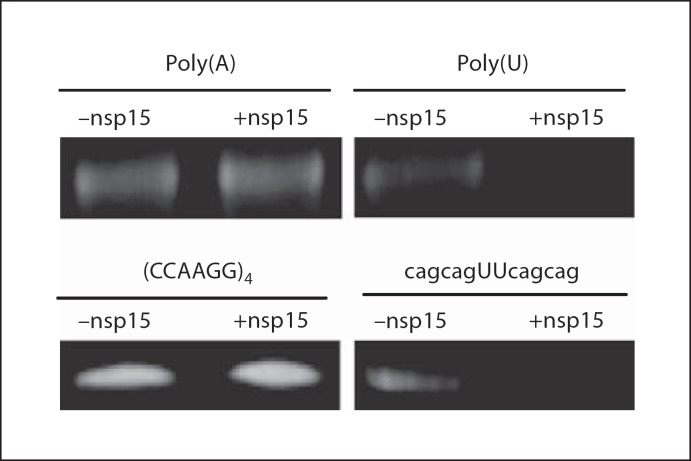
figure 3 shows that TCoV nsp15 quickly degraded poly(U)50 but did not degrade poly(A)50. figure 3 also shows that nsp15 did not degrade ssRNA oligo TJ1 which contained no U in the sequence. However, nsp15 degraded TJ2, which contained UU in the middle of the sequence. Collectively, these data clearly demonstrated that nsp15 cleaves U-containing ssRNA with poly(U) as the preferable substrate.
Fig. 3.
Substrate specificity of TCoV nsp15. Synthetic ssRNA oligo poly(A)50, poly(U)50, (CCAAGG)4, and cagcagUUcagcag were incubated in the presence or absence of purified TCoV nsp15 for 30 min at 37° and then separated on denaturing PAGE.
TCoV nsp15 Cleavage of dsRNA
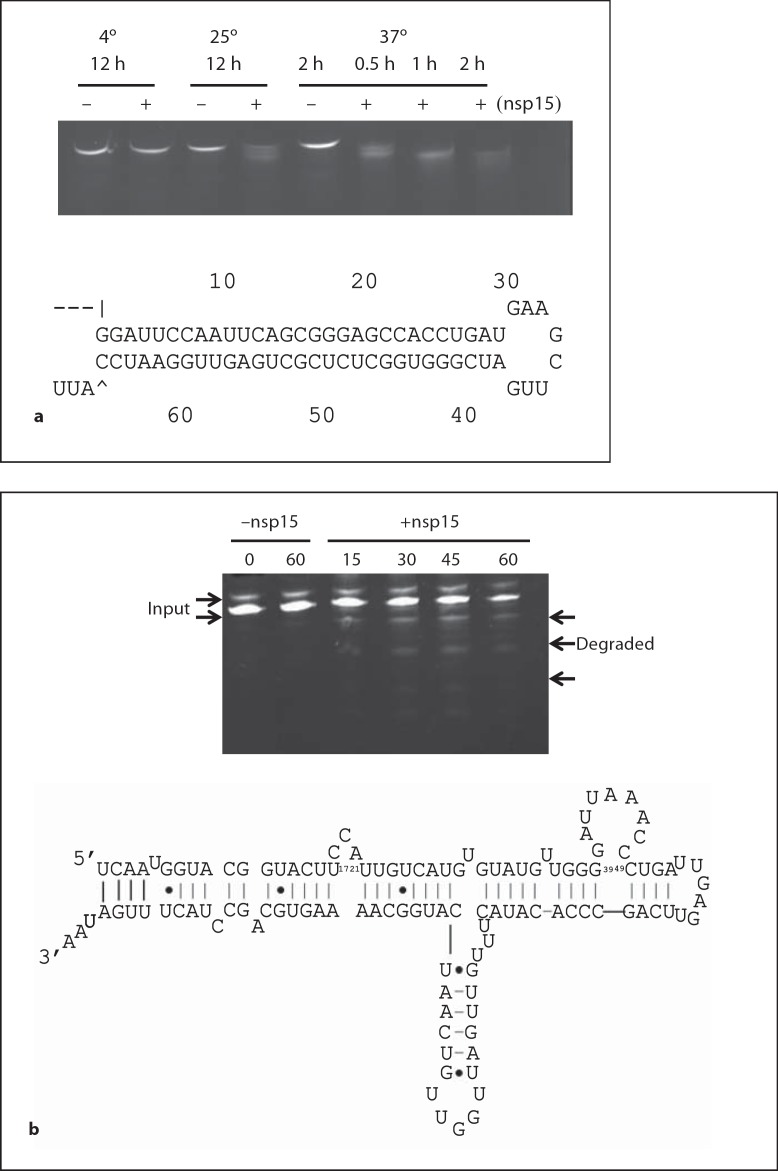
figure 4a shows degradation of hairpin RNA by nsp15. The hairpin RNA had a dsRNA region of 29-nt and a loop of 8-nt in the middle of the sequence and extra 3-nt at the 3′ end. There were 14 potential cuts for nsp15 within the dsRNA region and two potential cuts within the loop region. When the reaction was incubated at 4° for 12 h, there was almost no degradation of the RNA. When the incubation temperature was increased to 25°, two degraded bands appeared, indicating some degradation. The band sizes suggested that the RNA was cut at the loop region to generate dsRNA that separated into two different-sized ssRNA bands on the gel. When the incubation temperature was increased to 37°, the degradation rate increased greatly. Within 30 min, the input RNA was degraded and two product bands appeared. After 60 min, there was only one band detected. Two hours later, the band was weak and a smear appeared, suggesting the RNA was cleaved at the loop region containing the unpaired U. Although there were several Us within the dsRNA region, cleavage at these positions seemed unlikely.
Fig. 4.
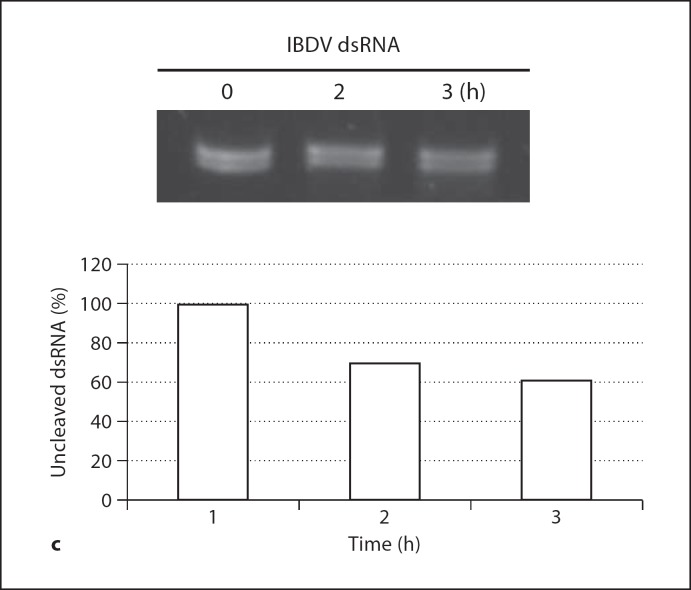
1 Cleavage of hairpin RNA by TCoV nsp15. Hairpin RNA was incubated with purified nsp15 for different times at different temperatures and then separated on denaturing PAGE. b Cleavage of pRNA by TCoV nsp15. pRNA was incubated with purified nsp15 for different amount of time at 37° and separated on denaturing PAGE. The arrows indicate the degraded products. The lower panel displays the sequence and predicted 2° structure of the pRNA. Fig. 4. c Cleavage of IBDV genomic dsRNA by TCoV nsp15. IBDV genomic dsRNA was isolated from infected bursa (see Materials and Methods) and incubated with 100 ng TCoV nsp15 at 37° for the indicated time. RNA was separated on 1.2% agarose gel and undigested RNA was measured by densitometry (displayed in the lower figure). More than 60% of input RNA remained undigested after 3 h of incubation.
When pRNA containing several dsRNA and loop regions was used as a substrate, nsp15 cleaved pRNA slowly and at specific positions to give a fixed pattern on the gel from 15 to 60 min of incubation (fig. 4b). figure 4c shows degradation of IBDV dsRNA by TCoV nsp15. After 3 h incubation, more than 60% of the input RNA remained undigested, indicating that dsRNA was not a good substrate for nsp15. All of these results suggest that nsp15 cleaves the loop regions within dsRNA, but cleavage in double-stranded regions is unlikely.
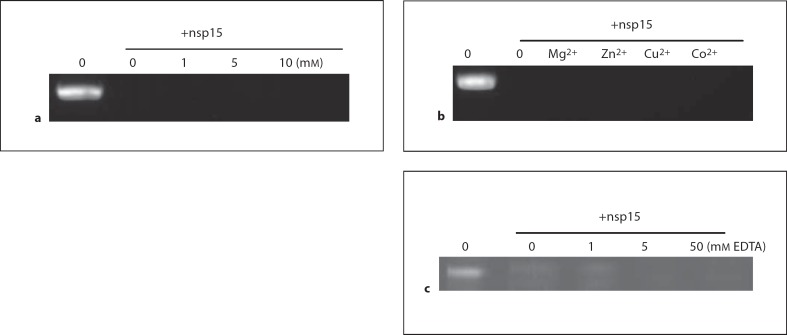
Requirement of Metal Ions for TCoV nsp15 Activity
figure 5a shows that in the absence of added Mn2+, nsp15 cleaves ssRNA as quickly as in its presence, raising the question of whether divalent metal ions are required for nsp15 RNase activity. figure 5b showed that several other divalent ions (Cu2+, Zn2+, Co2+, Mg2+) also did not affect the RNase activity of nsp15. To further examine the role of divalent metal ions in nsp15 activity, nsp15 was preincubated with different concentrations of EDTA, a chelating agent for divalent metal ions, to determine whether the EDTA could inhibit nsp15 activity by chelating the divalent ions. figure 5c indicates that degradation of ssRNA by nsp15 is not inhibited by preincubation of the nsp15 with the EDTA up to 50 mM. Rather, higher EDTA concentrations seemed to increase the rate of RNA cleavage by TCoV nsp15.
Fig. 5. a.
Effect of Mn2+ on activity of nsp15. nsp15 was incubated with different concentrations of Mn2+ for 10 min on ice and then RNA was added and reaction was incubated for 60 min at 37°. b Effect of divalent metal ions on nsp15 activity. nsp15 was incubated with 5 mM of different ions for 10 min on ice and then RNA was added and reaction was incubated for 60 min at 37°. c Effect of preincubation of nsp15 with EDTA on nsp15 activity. Purified TCoV nsp15 was incubated with 0, 1, 5, and 50 mM EDTA for 10 min on ice and then RNA substrate was added and the reaction was incubated for 30 min at 37°.
Active Form of TCoV nsp15
Because TCoV nsp15 displayed different forms on SDS-PAGE in the absence of β-ME (data not shown), we explored which form(s) has the RNase activity. figure 6 shows the results of RNA zymography. When nsp15 was incubated in the absence of β-ME, only one band was visualized on zymography. The size of the band was around 250 kDa, corresponding to the size of a hexamer. However, there was no RNase activity when nsp15 was incubated in the presence of β-ME. These data strongly suggest that the only form that shows detectable RNase activity in vitro is a homohexamer, and both intra- or intermolecular disulfide bonds are necessary for the RNase activity of nsp15.
Fig. 6.
Zymography for TCoV nsp15. Purified TCoV nsp15 was incubated for 5 min in the presence (+) or absence (-) of 5% β-ME at 95° and then separated on 10% SDS-PAGE containing 0.1 mg/ml of yeast RNA. A band around 250 kDa was visualized in the absence of β-ME, corresponding to the size of a hexamer. See Materials and Methods for detailed procedure.
Discussion
Coronavirus polyprotein is a large protein that is processed into 15–16 functional nsps. The biological relevance of these nsps is not clear, but they are believed to be involved in RNA genome replication and transcription. We predicted that there are 8 enzyme domains within the polyprotein 1ab of TCoV, including 3-CLpro, PLP, RdRp, and helicase [20]. Within coronavirus, the in vitro activities of the other four enzymes were recently reported. SARS-CoV nsp14 showed 3′ to 5′ exoribonuclease activity [21], and a single mutation Tyr6398His in mouse hepatitis virus MHV-A59 nsp14 resulted in attenuated virus pathogenesis in mice [22]. HCoV-229E ADP-ribose1′-monophosphate (Appr-1′-p) processing enzyme was demonstrated to dephosphorylate Appr-1′-p [23]. The most extensively investigated enzyme was NendoU, whose in vitro activity was demonstrated in SARS-CoV [10, 24], HCoV-229E [11], MHV [12], and EAV [13].
The TCoV nsp15 shares very high sequence identity with those of other coronavirus NendoUs (fig. 1a), and the predicted key residues associated with enzymatic activity are conserved. Using bacterial-expressed recombinant nsp15, we clearly demonstrated that the TCoV nsp15 is a NendoU. First, the TCoV nsp15 cleaved total cellular RNA randomly but did not cleave ssDNA or dsDNA, demonstrating that TCoV nsp15 is an RNase. Second, the TCoV nsp15 did not cleave single-stranded poly(A) RNA or the synthetic ssRNA TJ1 that did not contain any U within the sequence. Third, the TCoV nsp15 cleaved poly(U) and U-containing ssRNA TJ2 efficiently. This evidences strongly supports our hypothesis that TCoV nsp15 is an endoribonuclease that preferentially cleaves U-containing RNA and is thus a NendoU.
While nsp15 from SARS-CoV, MHV, and HCoV-229E cleaved dsRNA as efficiently as ssRNA, nsp15 of TCoV did not (fig. 4). With respect to dsRNA of IBDV, hairpin RNA, and pRNA, TCoV nsp15 cleaved these dsRNA molecules very slowly. In particular, TCoV nsp15 cleavage of pRNA yielded a fixed pattern on the gel, suggesting that TCoV nsp15 did not cleave randomly, but at specific sites. The same was true for hairpin RNA. There was no smear on the gel as compared with smears for total cellular RNA (fig. 2). Whether or not this difference in cleaving dsRNA between nsp15 of TCoV and other coronaviruses has some biological significance require more detailed investigation.
The TCoV nsp15 did not require Mn2+ or any other divalent metal ions for its enzymatic activity (fig. 6), a characteristic of XendoU and NendoU in SARS-CoV and HCoV-229E. Preincubation of the TCoV nsp15 with increasing concentrations of EDTA slightly increased its activity (fig. 5c). This observation was in contrast to NendoU of SARS-CoV, HCoV-229E, MHV, and EAV [10, 13, 25]. Whether this difference is rudimental or has biological relevance is not yet clear. Because TCoV is a group 3 coronavirus, and nsp15 of TCoV and IBV share over 90% sequence similarity, it is worth determining whether or not IBV nsp15 requires Mn2+ as a cofactor. Recent studies on the crystal structures of NendoU of SARS-CoV and MHV failed to demonstrate coordinating package of Mn2+ within the crystal [12]. One possibility is that Mn2+ enhances affinity between nsp15 and the RNA molecule. Recent data from SARS-CoV showing that Mn2+ increased interaction between a SARS nsp15 mutant protein, H234A, and a synthetic RNA, rU16 [24], are consistent with our hypothesis.
RNA zymography demonstrated that the active form of the TCoV nsp15 is a hexamer. This was consistent with results obtained for SARS and MHV NendoU [12, 14]. Our data also revealed for the first time that disulfide bonds are involved in the formation of a trimer or hexamer and are essential for the enzymatic activity of the TCoV nsp15 (fig. 6). The monomer or trimer showed no RNase activity. Intramolecular disulfide bonds within the TCoV nsp15 were observed. The hexamer form of NendoU was demonstrated in SARS and MHV NendoU [12, 14]. Recent crystal structure analysis revealed that the hexamer of NendoU of SARS-CoV and MHV was formed by two trimers through back-to-back interactions [12, 16]. Our zymography results about the TCoV nsp15 as well as data from other NendoU indicate the in vitro active form of the NendoU is a homohexamer; whether this is the case in vivo requires further investigation.
The biological function of NendoU is still a mystery. Using reverse genetics, a point mutation in HCoV-229E NendoU resulted in a lack of viral genome replication and transcription [11]. However, no rescue experiment was performed to demonstrate that it was the RNase activity of the enzyme which caused the failure in genome replication and transcription. In EAV, extensive site-directed mutagenesis experiments revealed that the loss of the whole NendoU was lethal for the virus, while point mutations of the enzyme mostly affected the production of sgRNA [13]. Considering that the coronavirus genome is ssRNA, having two RNases (nsp14 and nsp15) in the genome seems peculiar for the virus. For the RNases to exert their functions but not degrade viral genomic RNA and sgRNA, the activity and specificity of the RNases must be under tight control. Therefore, it is expected that viral or cellular proteins play roles in regulating NendoU activity and specificity.
In conclusion, we have demonstrated that nsp15 of TCoV is a novel NendoU which only cleaves U-containing ssRNA in the absence of Mn2+. The in vitro active form of TCoV nsp15 is a hexamer, and disulfide bonds are necessary for hexamer formation and enzyme activity.
References
- 1.Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai MM. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 4.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziebuhr J. The coronavirus replicase. Curr Top Microbiol Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasternak AO, Spaan WJ, Snijder EJ. Nidovirus transcription: how to make sense..? J Gen Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 7.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laneve P, Altieri F, Fiori ME, Scaloni A, Bozzoni I, Caffarelli E. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J Biol Chem. 2003;278:13026–13032. doi: 10.1074/jbc.M211937200. [DOI] [PubMed] [Google Scholar]
- 10.Bhardwaj K, Guarino L, Kao CC. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Zhai Y, Sun F, Lou Z, Su D, Xu Y, Zhang R, Joachimiak A, Zhang XC, Bartlam M, Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posthuma CC, Nedialkova DD, Zevenhoven-Dobbe JC, Blokhuis JH, Gorbalenya AE, Snijder EJ. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J Virol. 2006;80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarino LA, Bhardwaj K, Dong W, Sun J, Holzenburg A, Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J Mol Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricagno S, Coutard B, Grisel S, Bremond N, Dalle K, Tocque F, Campanacci V, Lichiere J, Lantez V, Debarnot C, Cambillau C, Canard B, Egloff MP. Crystallization and preliminary X-ray diffraction analysis of Nsp15 from SARS coronavirus. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2006;62:409–411. doi: 10.1107/S1744309106009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricagno S, Egloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshmukh DR, Sautter JH, Patel BL, Pomeroy BS. Histopathology of fasting and bluecomb disease in turkey poults and embryos experimentally infected with bluecomb disease coronavirus. Avian Dis. 1976;20:631–640. [PubMed] [Google Scholar]
- 18.Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- 19.Saif LJ. Coronavirus immunogens. Vet Microbiol. 1993;37:285–297. doi: 10.1016/0378-1135(93)90030-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao J, Wu CC, Lin TL. Complete nucleotide sequence of polyprotein gene 1 and genome organization of turkey coronavirus. Virus Res. 2008;136:43–49. doi: 10.1016/j.virusres.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperry SM, Kazi L, Graham RL, Baric RS, Weiss SR, Denison MR. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J Virol. 2005;79:3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putics A, Filipowicz W, Hall J, Gorbalenya AE, Ziebuhr J. ADP-ribose-1′-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2006;361:243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang H, Bhardwaj K, Li Y, Palaninathan S, Sacchettini J, Guarino L, Leibowitz JL, Kao CC. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]