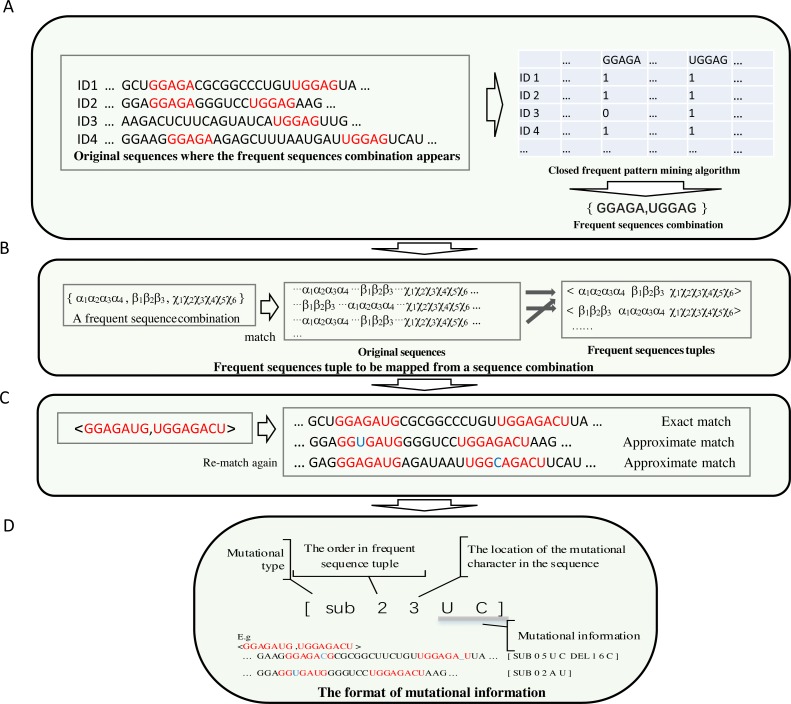
Figure 2. The demonstration of the algorithm in detail.
(A) Each frequent sequence maps to a column of a table. Each sub-sequence is represented by a symbol, and the frequent sequences combinations were derived. (B) The order of each sub-sequences in the combination of each frequent sequence is obtained by re-scanning the original sequences. (C) The achievement of the frequent sequences approximate matching (implemented by non-deterministic finite automaton algorithm) is shown. (D) Compact data structure for recording mutation information. The first digit represents the type of mutation, which can be either sub(substitution), ins (insertion) or del (deletion). The number for location begins from zero.