Addition of sodium bicarbonate (NaHCO3) to standard antimicrobial susceptibility testing medium reveals certain methicillin-resistant Staphylococcus aureus (MRSA) strains to be highly susceptible to β-lactams. We investigated the prevalence of this phenotype (NaHCO3 responsiveness) to two β-lactams among 58 clinical MRSA bloodstream isolates. Of note, ∼75% and ∼36% of isolates displayed the NaHCO3 responsiveness phenotype to cefazolin (CFZ) and oxacillin (OXA), respectively.
KEYWORDS: antimicrobial susceptibility testing, beta-lactams, methicillin-resistant Staphylococcus aureus, sodium bicarbonate
ABSTRACT
Addition of sodium bicarbonate (NaHCO3) to standard antimicrobial susceptibility testing medium reveals certain methicillin-resistant Staphylococcus aureus (MRSA) strains to be highly susceptible to β-lactams. We investigated the prevalence of this phenotype (NaHCO3 responsiveness) to two β-lactams among 58 clinical MRSA bloodstream isolates. Of note, ∼75% and ∼36% of isolates displayed the NaHCO3 responsiveness phenotype to cefazolin (CFZ) and oxacillin (OXA), respectively. Neither intrinsic β-lactam MICs in standard Mueller-Hinton broth (MHB) nor population analysis profiles were predictive of this phenotype. Several genotypic markers (clonal complex 8 [CC8]; agr I and spa t008) were associated with NaHCO3 responsiveness for OXA.
INTRODUCTION
Staphylococcus aureus is a serious community and nosocomial pathogen and a leading cause of bacteremia, infective endocarditis, and device-related infections (1, 2). Many of these infections are caused by methicillin-resistant S. aureus (MRSA), which is generally perceived to be “resistant” to those β-lactam antibiotic therapies used for the treatment of methicillin-susceptible S. aureus (MSSA) (3). MRSA exhibits in vitro resistance to oxacillin (OXA) by standard antimicrobial susceptibility testing (AST), which has been extrapolated to apply to all other β-lactams (excluding ceftaroline and ceftobiprole) (4). However, as shown recently, in vitro antimicrobial resistance of MRSA (and selected Gram-negative bacilli) may not always correlate to therapeutic resistance in vivo (5–8). Thus, efforts have been made to more effectively model the host environment in newer in vitro AST to improve the predictive power of these assays with regard to clinical outcomes (6, 7, 9). These modified AST protocols utilize tissue culture media or media that contain specific ions that a pathogen would consistently encounter within a host (e.g., sodium bicarbonate [NaHCO3]).
Recently, we identified a novel MRSA AST phenotype, termed “NaHCO3 responsiveness,” in which MRSA strains are highly susceptible in vitro to cefazolin (CFZ) and OXA in a medium supplemented with NaHCO3 (7). Prototype MRSA strains with this in vitro phenotype were also highly susceptible to these same β-lactams in an ex vivo simulated endocarditis vegetation model as well as in a rabbit model of infective endocarditis (7, 10). In contrast, in vitro NaHCO3-nonresponsive isolates were also resistant to CFZ and OXA under these same ex vivo and in vivo conditions.
The aim of this study was to delineate the frequency of the NaHCO3-responsive/nonresponsive phenotypes in vitro among a larger MRSA strain set. Thus, we analyzed a collection of 58 MRSA bloodstream isolates for the following to determine potential predictive markers of NaHCO3 responsiveness: (i) NaHCO3-responsive phenotypes to CFZ and OXA and (ii) linkage of this phenotype with other known phenotypic and genotypic markers. We identified a relatively large subset of NaHCO3-responsive strains to CFZ and/or OXA within this MRSA cohort.
(This study was presented in part at the IDWeek Conference, Washington, DC, October 2019 [11].)
Frequency of NaHCO3 responsiveness.
Among 58 clinical MRSA isolates, 22 were vancomycin- and daptomycin-susceptible bloodstream isolates obtained from the Cubist Isolate Collection. The remaining 36 strains (also vancomycin and daptomycin susceptible) were from bacteremic patients at Duke University Medical Center (kindly provided by Vance Fowler) (12). Fifty-five of the 58 isolates were β-lactamase positive as determined by nitrocefin disk assay as per manufacturer’s instructions (Becton, Dickinson).
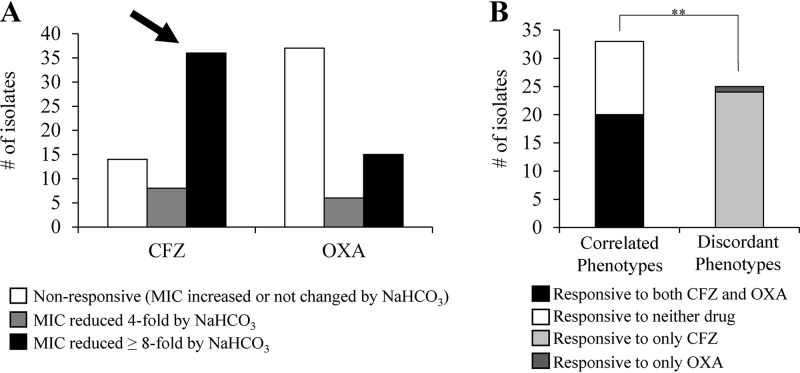
The NaHCO3-responsive phenotype to CFZ and OXA was defined as ≥4-fold reduction in MICs in the presence versus absence of 44 mM NaHCO3 by broth microdilution assay (7). Previously, we determined that this in vitro criterion was a good predictor of in vivo outcomes in a rabbit model of MRSA infective endocarditis (7). The majority of strains displayed NaHCO3 responsiveness to at least one of the two β-lactams (Fig. 1A; see also Table S1 in the supplemental material). Thus, 44/58 (76%) and 21/58 (36%) tested strains displayed reduced MICs to CFZ and OXA, respectively, in the presence of NaHCO3 (Fig. 1A). Additionally, the majority of such responsive strains displayed a ≥8-fold reduction in MICs in the presence of NaHCO3 (82% for CFZ and 71% for OXA) (Fig. 1A).
FIG 1.
Frequency of NaHCO3 responsiveness among 58 clinical MRSA isolates. (A) Frequency of NaHCO3 responsiveness to cefazolin (CFZ) and oxacillin (OXA). Responsiveness is defined as a ≥4-fold reduction in MIC in the presence of NaHCO3 compared to that of medium lacking NaHCO3. (B) Frequency of coresponsiveness to CFZ and OXA. Coresponsiveness is defined as a strain that is NaHCO3 responsive to both CFZ and OXA. Correlated phenotypes are those in which a strain is either responsive to both drugs (coresponsive) or responsive to neither drug (nonresponsive). Discordant phenotypes are those in which a strain is responsive to only one drug (either CFZ or OXA). Kappa coefficient of correlation (κ) = 0.25, **, P = 0.008.
We identified 20/58 (35%) strains to be coresponsive to both CFZ and OXA, while 13/58 (22%) strains were responsive to neither drug (Fig. 1B). The significance of this correlation was calculated by the kappa coefficient (κ) for the coresponsive phenotype. Although the kappa coefficient indicated a positive correlation for this coresponsiveness, there were also a significant number of strains with discordant phenotypes (NaHCO3 responsive to only CFZ or only OXA) in our cohort sample (McNemar’s test; ****, P < 0.0001) (Fig. 1B). The majority of coresponsive phenotypes appeared to be driven by the 20/21 OXA-responsive strains that were also responsive to CFZ. These data indicate that testing for NaHCO3 responsiveness to OXA is a reasonable proxy for NaHCO3 responsiveness to CFZ but not vice versa.
Recent interest has focused on the use of the tissue culture medium, RPMI 1640, for AST as a more host-mimicking milieu, particularly for Gram-negative bacteria (13–15). In a previous study of five prototype MRSA strains, we determined that AST in RMPI 1640 (containing ∼25 mM NaHCO3) resulted in a ≥4-fold decrease in MICs to CFZ and OXA, despite only two of the five becoming susceptible to these same β-lactams in 44 mM NaHCO3-Mueller-Hinton broth (MHB) medium. Moreover, only the two NaHCO3-responsive strains were cleared in vivo from experimental endocarditis target tissues by these two β-lactams (7). To further assess the impact of RPMI medium on MRSA β-lactam MICs in the current cohort, 58/58 (100%) and 50/58 (86%) strains were rendered highly susceptible to CFZ and OXA, respectively, when tested in RMPI 1640 (see Table S1). Thus, RPMI medium does not appear to be a discriminative candidate as a host-mimicking medium for new MRSA AST.
Population analysis profiles, time-kill synergy, and intrinsic (baseline) MICs of NaHCO3-responsive versus nonresponsive strains.
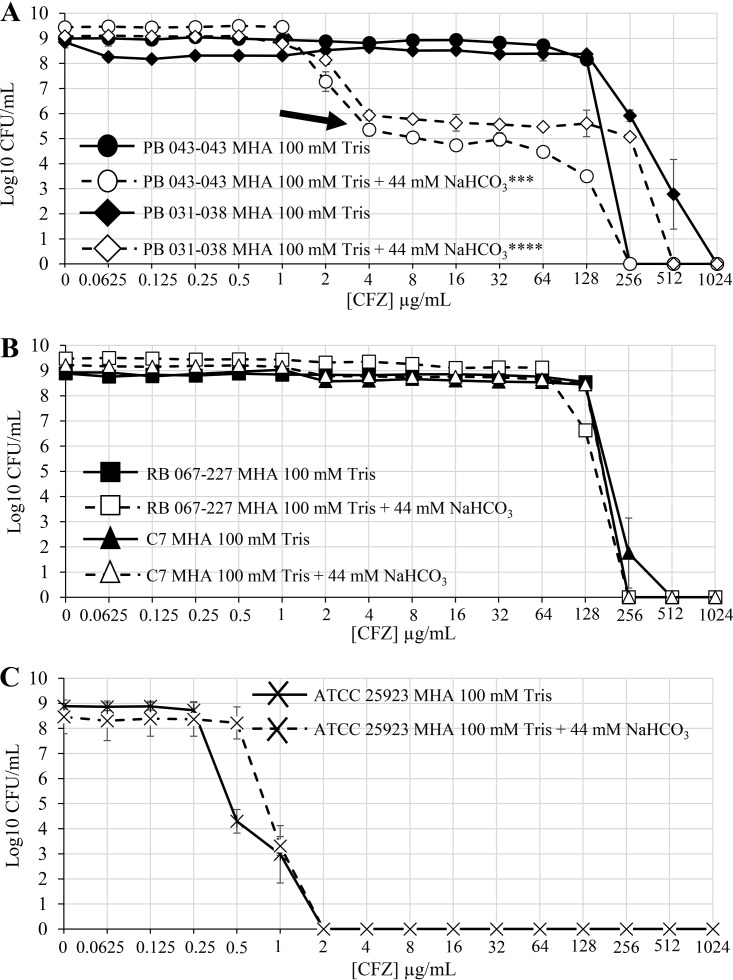
Homoresistant MRSA strains are typically classified as those in which essentially the entire cell population displays the same level of resistance to an antibiotic of interest by population analysis profile (PAP) (16). In contrast, in heteroresistant strains, only a relatively small “subpopulation” of cells within the entire population display high-level resistance. Thus, PAPs were carried out on two responsive (PB 043-043 and PB 031-038) and two nonresponsive MRSA strains (C7 and RB 067-227) from our cohort in the presence and absence of NaHCO3 as previously described (7). All four strains exhibited a homoresistant CFZ PAP in standard Mueller-Hinton agar (MHA). Of note, both responsive strains demonstrated a significant repression of the overall resistant subpopulation in the presence of NaHCO3 (Fig. 2A). In contrast, neither nonresponsive strain PAP was impacted by growth in NaHCO3-containing medium (Fig. 2B). As expected, the PAP of an MSSA isolate determined in medium ± NaHCO3 was homogenously susceptible to CFZ in both conditions (Fig. 2C). Upon further PAP testing of the remaining strains within the cohort, a large proportion of responsive strains had homoresistant PAPs, with only two strains exhibiting heteroresistant PAPs (see Table S2 in the supplemental material). Similarly, the vast majority of nonresponsive strains tested had homoresistant PAPs, but this was not uniformly found among this group (Table S2). Thus, there was no obvious linkage between NaHCO3 responsiveness and heteroresistance (7) or NaHCO3 nonresponsiveness and homoresistance. These data underscore the notion that NaHCO3 responsiveness/nonresponsiveness is not merely a reflection of the proportion of the β-lactam-resistant subpopulations within a given MRSA strain; it is more likely that complex microbial factors are in-play.
FIG 2.
Population analysis profiles for NaHCO3-responsive and nonresponsive strains. (A) Population analysis profiles of CFZ for NaHCO3-responsive strains in the presence and absence of NaHCO3. The area under the curve (AUC), calculated by linear approximation, was significantly decreased by exposure to NaHCO3 in both strains as analyzed by Student’s t test (PB 043-043, ***, P < 0.001; PB 031-038, ****, P < 0.0001). (B) Population analysis profiles of CFZ for NaHCO3-nonresponsive strains in the presence and absence of NaHCO3. As analyzed by Student’s t test, there is no significant difference in the AUC for C7 in media ± NaHCO3; the AUC of RB 067-227 was significantly increased by exposure to NaHCO3 (*, P < 0.05). (C) Population analysis profiles of CFZ for the methicillin-susceptible S. aureus (MSSA) strain ATCC 25923.
We assessed potential synergistic killing between NaHCO3 plus β-lactams in responsive versus nonresponsive strains by time-kill assay (7). Synergy was defined as a ≥2 log10 CFU/ml difference in counts at 24 h when comparing the β-lactam agent alone versus the combination of β-lactam plus NaHCO3. To qualify for synergy, there also had to be a bactericidal impact of the β-lactam plus NaHCO3 combination (i.e., ≥3 log10 CFU/ml decrease from the 0 h starting inoculum). In two responsive strains, only one strain (PB 043-043) exhibited both a bactericidal and synergistic impact of CFZ plus NaHCO3 (Fig. 3A and B). In contrast, neither nonresponsive strain displayed either a bactericidal or synergistic impact of CFZ or OXA plus NaHCO3 versus medium alone (Fig. 3C and D).
FIG 3.
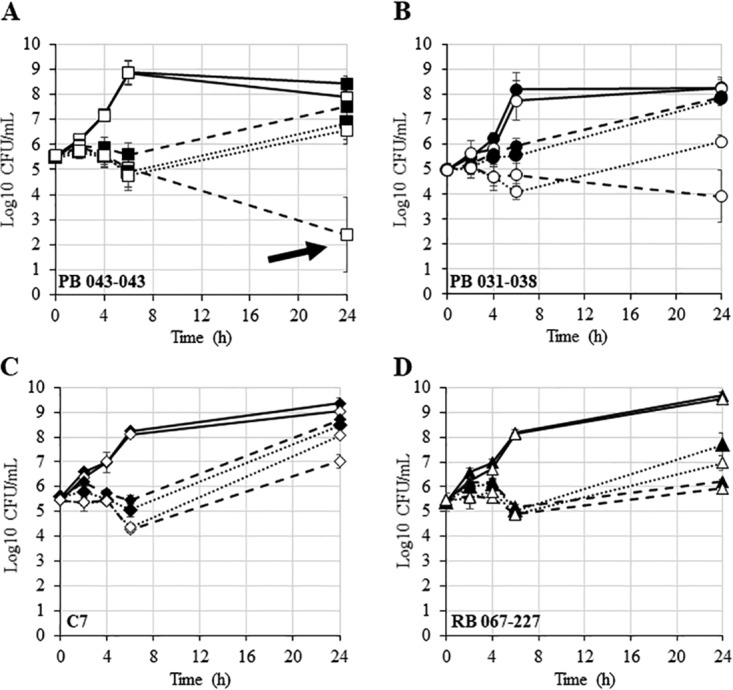
Time kill curves for NaHCO3-responsive and nonresponsive strains. (A) PB 043-043 (squares). (B) PB 031-038 (circles). (C) C7 (diamonds). (D) RB 067-227 (triangles). Closed symbols represent growth in CA-MHB 100 mM Tris, open symbols represent growth in CA-MHB 100 mM Tris + 44 mM NaHCO3, solid lines represent no drug control, dashed lines represent exposure to CFZ, and dotted lines represents exposure to OXA. Drug concentrations for PB 043-043 were 16 μg/ml for CFZ and 32 μg/ml for OXA; drug concentrations for all other strains were 64 μg/ml for both CFZ and OXA.
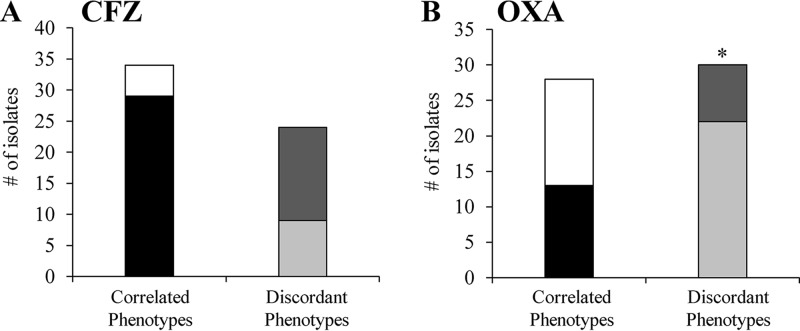
We assessed whether the NaHCO3-responsive phenotype might be predicted by a relatively low intrinsic (baseline) MIC to CFZ and OXA in standard MHB (i.e., an MIC in standard community-associated methicillin-resistant Staphylococcus aureus [CA-MHB] of <64 μg/ml). However, such lower intrinsic MICs were only randomly associated with NaHCO3 responsiveness for both drugs (Fig. 4A and B).
FIG 4.
Relationship between intrinsic MIC and responsiveness for CFZ and OXA. (A) CFZ. (B) OXA. Both graphs show low intrinsic MIC/responsive (black), high intrinsic MIC/nonresponsive (white), low intrinsic MIC/nonresponsive (light gray), and high intrinsic MIC/responsive (dark gray). Relationship between intrinsic MIC and responsiveness for CFZ is random (κ = 0.01; 95% confidence interval (CI) = −0.24 to 0.27; Kappa coefficient and McNemar’s test, P > 0.05). Relationship between intrinsic MIC and responsiveness to OXA is also random with significant discordance (κ = 0.02; 95% CI = −0.21 to 0.25; Kappa coefficient, P > 0.05; McNemar’s test *, P = 0.01).
Taken together, the above data indicate that other intrinsic microbial properties, rather than the proportion of resistant cells within a population or the magnitude of that resistance, dictate the net bacteriostatic and/or bactericidal effects of CFZ and OXA in the presence of NaHCO3.
Genotypic correlates of NaHCO3 responsivity.
We determined potential relationships between several well-characterized genotypic markers in MRSA with the NaHCO3-responsive phenotype for either β-lactam, employing clonal complex (CC) type, agr type, spa type, and staphylococcal cassette chromosome mec element (SCCmec) type. The CC, agr, spa, and SCCmec types were determined as previously described (17–20).
There was a broad range of CC types observed in our cohort (see Table S1); however, only CC types 5 and 8 provided a large enough sample size to make any statistical inferences. Neither of these two CC types was a statistically better predictor of responsiveness to CFZ (see Fig. S1A in the supplemental material). Interestingly, CC8 strains had a significantly higher proportion of OXA-NaHCO3-responsive strains than CC5 strains (Fig. S1B). Similar to what was observed in the CC type data, neither agr type I or II strains were a better predictor of responsiveness to CFZ (see Table S1; chi-square analysis; P = 0.9); in contrast, agr type I strains exhibited a significantly higher frequency of OXA NaHCO3 responsiveness than agr type II strains (chi-square analysis; **, P = 0.009). Our cohort consisted of a diverse array of spa types (17 different variants; see Table S1); however, only spa types t008 and t002 were frequent enough to merit statistical analysis. As with CC and agr types, neither spa type was significantly linked to CFZ responsiveness, while spa type t008 was significantly linked to OXA responsiveness (Table S1). Although our statistical analyses indicate that certain genotypic markers are more commonly associated with OXA-NaHCO3 responsiveness, our sample size is too small to draw any definitive predictive conclusions.
The majority of strains (59%) were either SCCmec type IV (59%) or SCCmec type II (38%) (Table S1), with no significant associations with NaHCO3 responsivity for CFZ or OXA.
It appears that the NaHCO3 responsiveness phenotype and sensitization of MRSA to two prototype standard-of-care β-lactams is relatively frequent, especially for CFZ. This phenotype does not appear to be related to the size of the resistant subpopulation or the intrinsic MIC of responsive strains. Although our data suggest that certain genotypic markers may be linked to OXA responsiveness, a larger cohort is needed to verify this observation.
Interestingly, a recent study by Quach et al. identified a subset of closely related MRSA strains, falling within the multilocus sequence type ST8 grouping (a lineage encompassed within CC8), that display unexpected high rates of lysis in the presence of β-lactams (21). This may point to one potential mechanism contributing to NaHCO3 responsiveness, i.e., NaHCO3-enhanced, β-lactam-mediated lysis. We are currently examining this phenotype among the 58 MRSA isolates described in the current study.
In summary, if the association of in vitro NaHCO3 responsiveness and β-lactam sensitization can be further validated experimentally in vivo using larger MRSA cohorts, a well-designed clinical trial to verify this relationship may well be justified.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported in part by the following grants from the National Institutes of Health: 1R01-AI146078 (to A.S.B.) and 1R01-AI139244 (to Y.Q.X.).
We thank Brianne Zapata-Davila for technical assistance in these studies.
The authors declare that no individual conflicts of interest exist related to this manuscript.
All authors have read and approved the final manuscript. Specific contributions were as follows. Concept and design were performed by S.C.E., A.S.B., and Y.Q.X. Collection of data was performed by S.C.E., M.O., V.T.M., and J.M. Analysis of data was performed by S.C.E., Y.P., A.S.B., L.L., Y.Q.X., H.F.C., R.A.P., L.C., and B.K. Drafting the manuscript was completed by S.C.E., H.F.C., R.A.P., and A.S.B. Critical revision of the manuscript was completed by S.C.E., A.S.B., Y.Q.X., H.F.C., R.A.P., and V.G.F. All authors accepted the final manuscript and agreed to be accountable for all aspects of the work.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 2.Archer GL. 1998. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis 26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 3.Rayner C, Munckhof W. 2005. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern Med J 35:S3–S16. doi: 10.1111/j.1444-0903.2005.00976.x. [DOI] [PubMed] [Google Scholar]
- 4.Dien Bard J, Hindler JA, Gold HS, Limbago B. 2014. Rationale for eliminating Staphylococcus breakpoints for β-lactam agents other than penicillin, oxacillin or cefoxitin, and ceftaroline. Clin Infect Dis 58:1287–1296. doi: 10.1093/cid/ciu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler VG Jr, Sakoulas G, McIntyre LM, Meka VG, Arbeit RD, Cabell CH, Stryjewski ME, Eliopoulos GM, Reller LB, Corey GR, Jones T, Lucindo N, Yeaman MR, Bayer AS. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 190:1140–1149. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- 6.Ersoy SC, Heithoff DM, Barnes L, Tripp GK, House JK, Marth JD, Smith JW, Mahan MJ. 2017. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 20:173–181. doi: 10.1016/j.ebiom.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ersoy SC, Abdelhady W, Li L, Chambers HF, Xiong YQ, Bayer AS. 2019. Bicarbonate resensitization of methicillin-resistant Staphylococcus aureus to β-lactam antibiotics. Antimicrob Agents Chemother 63:e00496-19. doi: 10.1128/AAC.00496-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubicek-Sutherland JZ, Heithoff DM, Ersoy SC, Shimp WR, House JK, Marth JD, Smith JW, Mahan MJ. 2015. Host-dependent induction of transient antibiotic resistance: a prelude to treatment failure. EBioMedicine 2:1169–1178. doi: 10.1016/j.ebiom.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buyck JM, Plésiat P, Traore H, Vanderbist F, Tulkens PM, Van Bambeke F. 2012. Increased susceptibility of Pseudomonas aeruginosa to macrolides and ketolides in eukaryotic cell culture media and biological fluids due to decreased expression of oprM and increased outer-membrane permeability. Clin Infect Dis 55:534–542. doi: 10.1093/cid/cis473. [DOI] [PubMed] [Google Scholar]
- 10.Rose WE, Bienvenida AM, Xiong YQ, Chambers HF, Bayer AS, Ersoy SC. 16 December 2019. Ability of bicarbonate supplementation to sensitize selected methicillin-resistant Staphylococcus aureus (MRSA) strains to β-lactam antibiotics in an ex vivo simulated endocardial vegetation model. Antimicrob Agents Chemother doi: 10.1128/AAC.02072-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ersoy SC, Otmishi M, Milan VT, Li L, Pak Y, Mediavilla J, Chen L, Kreiswirth B, Chambers HF, Proctor RA, Xiong YQ, Fowler VG, Bayer AS. 2019. Abstr ID Week Conference, Washington, DC, October 2019, abstr 607. [DOI] [PMC free article] [PubMed]
- 12.Xiong YQ, Fowler VG Jr, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS. 2009. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J Infect Dis 199:201–208. doi: 10.1086/595738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin L, Nonejuie P, Munguia J, Hollands A, Olson J, Dam Q, Kumaraswamy M, Rivera H Jr, Corriden R, Rohde M, Hensler ME, Burkart MD, Pogliano J, Sakoulas G, Nizet V. 2015. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine 2:690–698. doi: 10.1016/j.ebiom.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumaraswamy M, Lin L, Olson J, Sun C-F, Nonejuie P, Corriden R, Döhrmann S, Ali SR, Amaro D, Rohde M, Pogliano J, Sakoulas G, Nizet V. 2016. Standard susceptibility testing overlooks potent azithromycin activity and cationic peptide synergy against MDR Stenotrophomonas maltophilia. J Antimicrob Chemother 71:1264–1269. doi: 10.1093/jac/dkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakoulas G, Rose W, Berti A, Olson J, Munguia J, Nonejuie P, Sakoulas E, Rybak MJ, Pogliano J, Nizet V. 2017. Classical β-lactamase inhibitors potentiate the activity of daptomycin against methicillin-resistant Staphylococcus aureus and colistin against Acinetobacter baumannii. Antimicrob Agents Chemother 61:e01745-16. doi: 10.1128/AAC.01745-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, Chambers HF. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob Agents Chemother 47:3040–3045. doi: 10.1128/aac.47.10.3040-3045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathema B, Mediavilla J, Kreiswirth BN. 2008. Sequence analysis of the variable number tandem repeat in Staphylococcus aureus protein A gene, p 285–305. In DeLeo FR, Otto M (eds), Bacterial pathogenesis. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 18.Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. 2012. Global epidemiology of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). Curr Opin Microbiol 15:588–595. doi: 10.1016/j.mib.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microbiol 69:18–23. doi: 10.1128/aem.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Mediavilla JR, Oliveira DC, Willey BM, de Lencastre H, Kreiswirth BN. 2009. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec typing. J Clin Microbiol 47:3692–3706. doi: 10.1128/JCM.00766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quach D, Sakoulas G, Nizet V, Pogliano J, Pogliano K. 2016. Bacterial cytological profiling (BCP) as a rapid and accurate antimicrobial susceptibility testing method for Staphylococcus aureus. EBioMedicine 4:95–103. doi: 10.1016/j.ebiom.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.