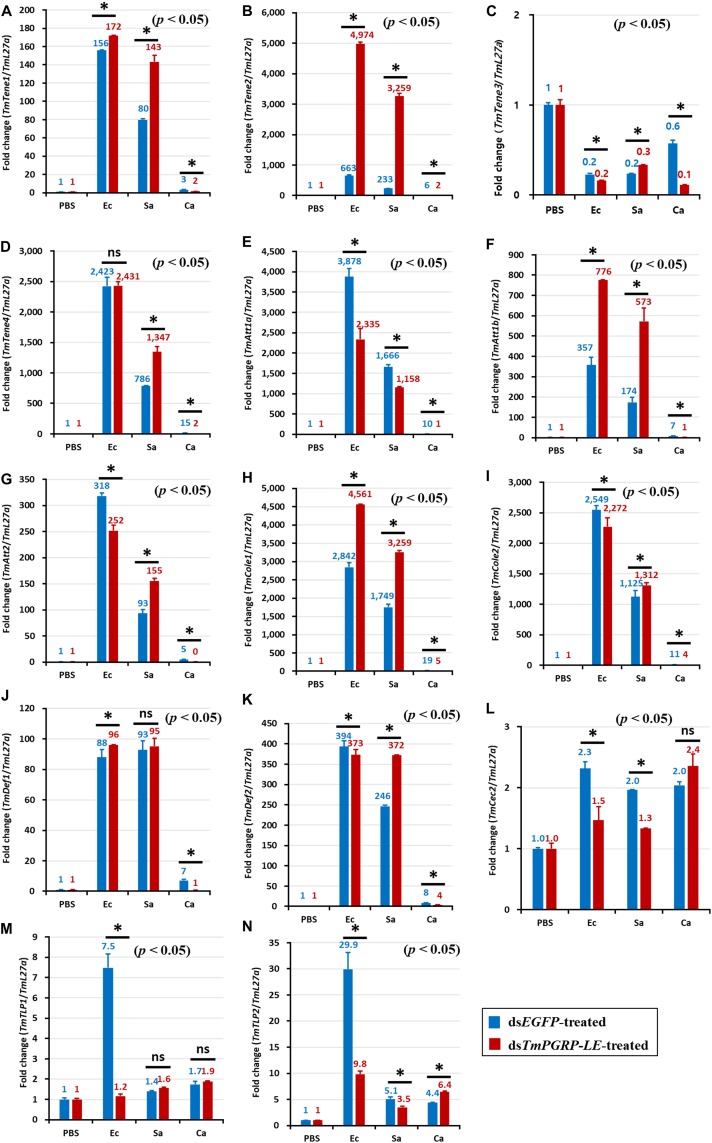
FIGURE 4.
Antimicrobial peptide (AMP) induction profiles in the TmPGRP-LE-silenced T. molitor larval fat body in response to E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) challenges. Double-stranded RNA specific to TmPGRP-LE was injected (1 μg) into young larvae. Six days after dsRNA treatment, TmPGRP-LE mRNA levels were reduced by 80% in the dsTmPGRP-LE-injected groups compared to dsEGFP-injected groups. The larvae were then infected with E. coli, S. aureus, and C. albicans (n = 20 per group). The expression levels of 14 AMP genes were evaluated by qRT-PCR: TmTenecin-1 (TmTene1, A); TmTenecin-2 (TmTene2, B); TmTenecin-3 (TmTene3, C); TmTenecin-4 (TmTene4, D); TmAttacin-1a (TmAtt1a, E); TmAttacin-1b (TmAtt1b, F); TmAttacin-2 (TmAtt2, G); TmColeptericin-1 (TmCole1, H); TmColeptericin-2 (TmCole2, I); TmDefensin-1 (TmDef1, J); TmDefensin-2 (TmDef2, K); TmCecropin-2 (TmCec2, L); TmTLP-1 (TmTLP1, M); and TmTLP-2 (TmTLP2, N). EGFP dsRNA injection served as a negative control, and the mRNA levels of the respective AMP genes are presented relative to those for TmL27a as an internal control. Each vertical bar represents mean ± SE of three independent biological replicates and the numbers above the bars show AMP transcription levels. Significant differences between dsEGFP- and dsTmPGRP-LE-treated cohorts are shown by asterisks (*) (p < 0.05) and ns, not significant.