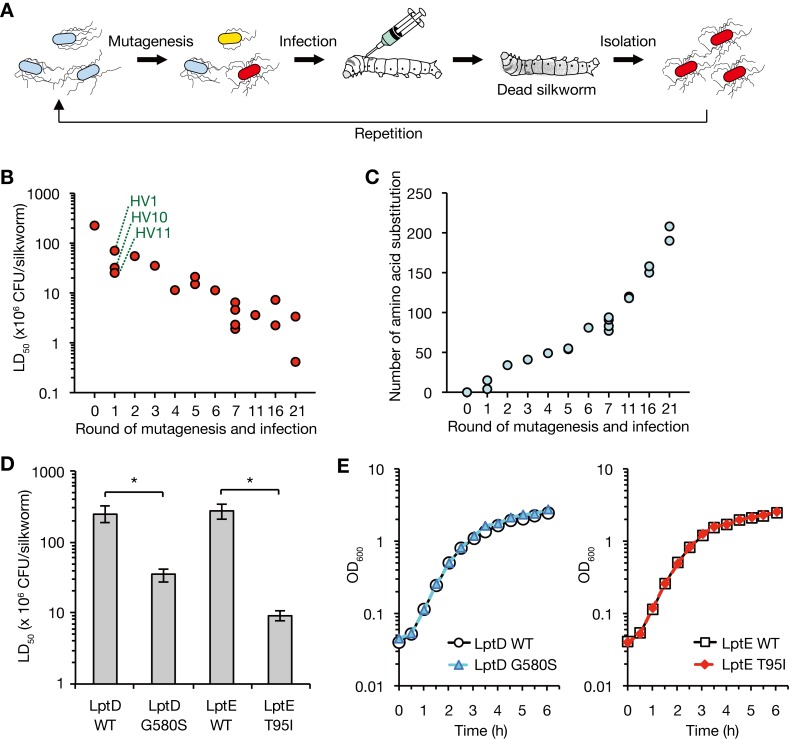
Fig 1. Isolation of E. coli mutants with high virulence by experimental evolution.
(A) A schematic representation of experimental evolution utilizing the silkworm infection model. E. coli strain treated with a mutagen was injected into silkworm. After the silkworm died, E. coli strains were isolated from the dead silkworm hemolymph. The mutagenesis and infection were repeated for 21 cycles. (B) An E. coli single colony isolated by the experimental evolution was cultured overnight, serially diluted, and injected into silkworms. Silkworm survival was determined at 48 h after the injection and the LD50 was determined by logistic regression. The horizontal axis represents the number of times the mutagenesis and infection experiments were performed. Each colony isolated from a dead silkworm that appeared different from the others was examined for its killing activity. The three symbols marked by green dotted lines represent the HV1, HV10, and HV11 strains. (C) E. coli strains isolated in the experimental evolution were subjected to whole genome sequencing and the number of amino acid substitutions is presented. The horizontal axis and strains are the same as in B. (D) Overnight cultured bacterial cells of the LptD WT, LptD G580S, LptE WT, and LptE T95I strains were serially diluted and injected into silkworms. Silkworm survival was determined at 48 h after the injection and the LD50 was determined by logistic regression. Data shown are the mean ± standard errors from three independent experiments. The asterisk represents a p value less than 0.05 (Student’s t test). (E) E. coli strains of LptD WT, LptD G580S, LptE WT, and LptE T95I were aerobically cultured in LB broth at 37˚C. The vertical axis represents the OD600 of bacterial culture, and the horizontal axis represents the culture time.