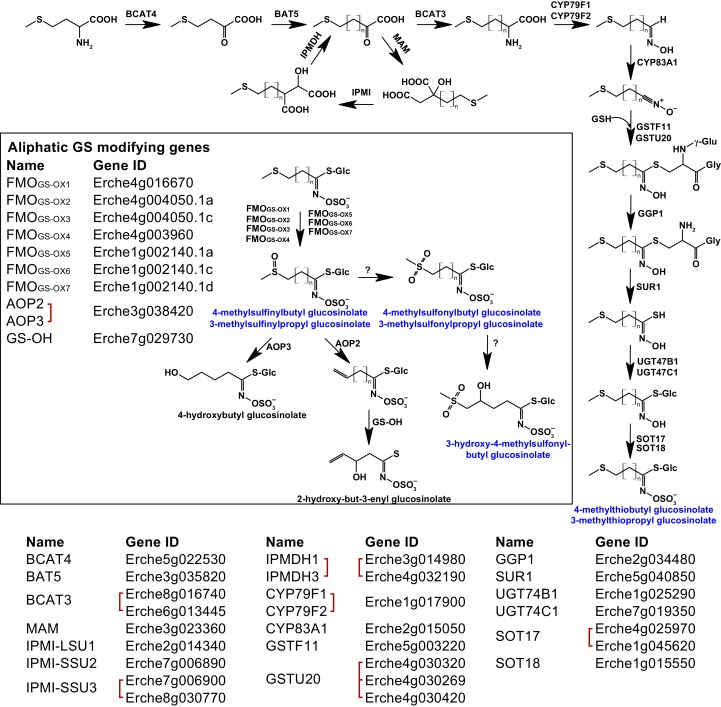
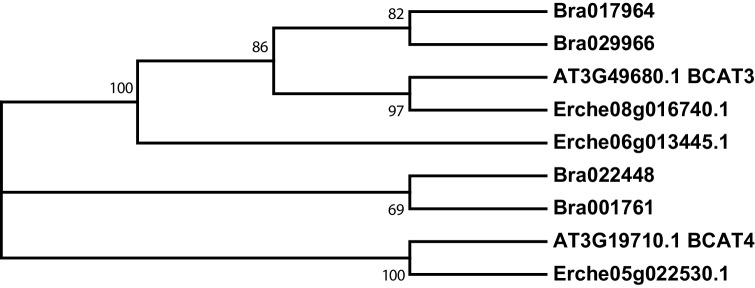
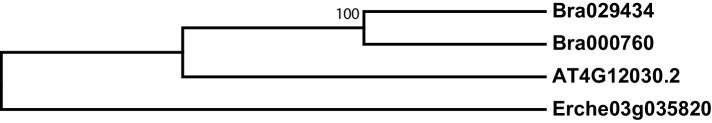
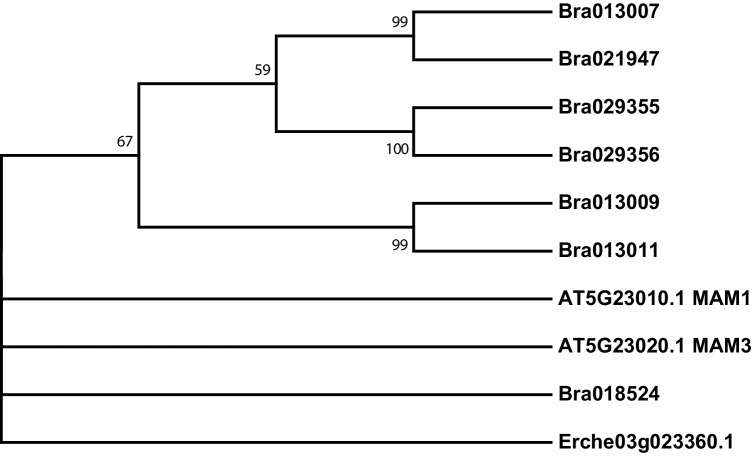
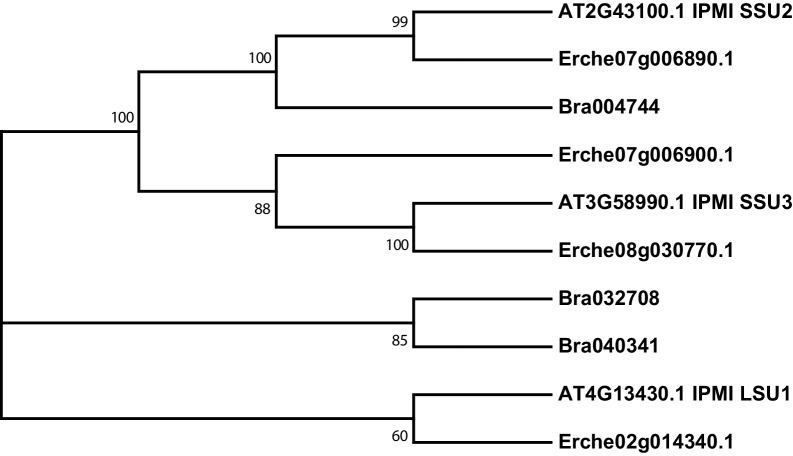
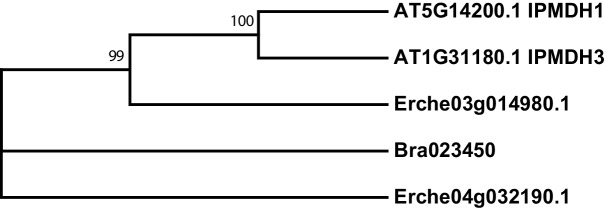
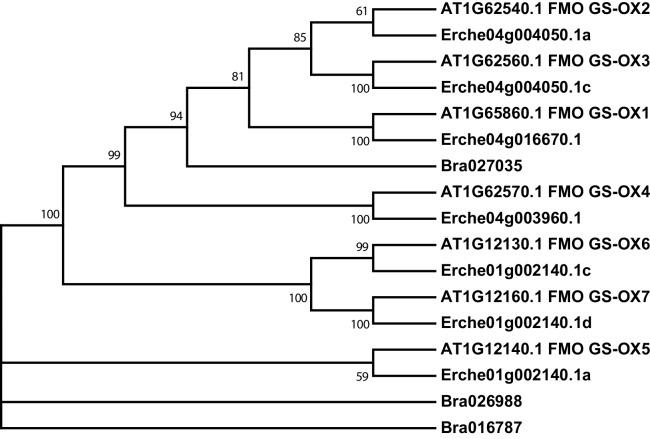
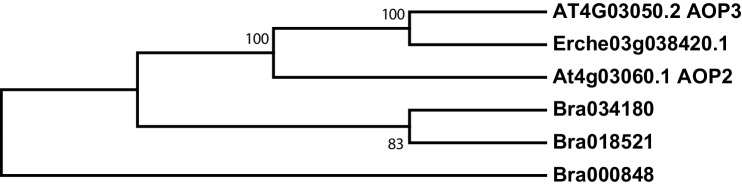
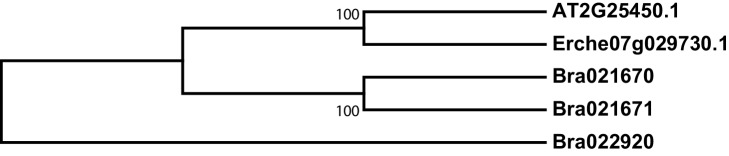
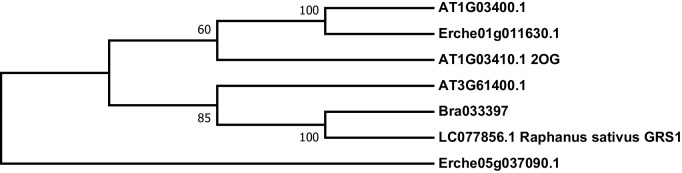
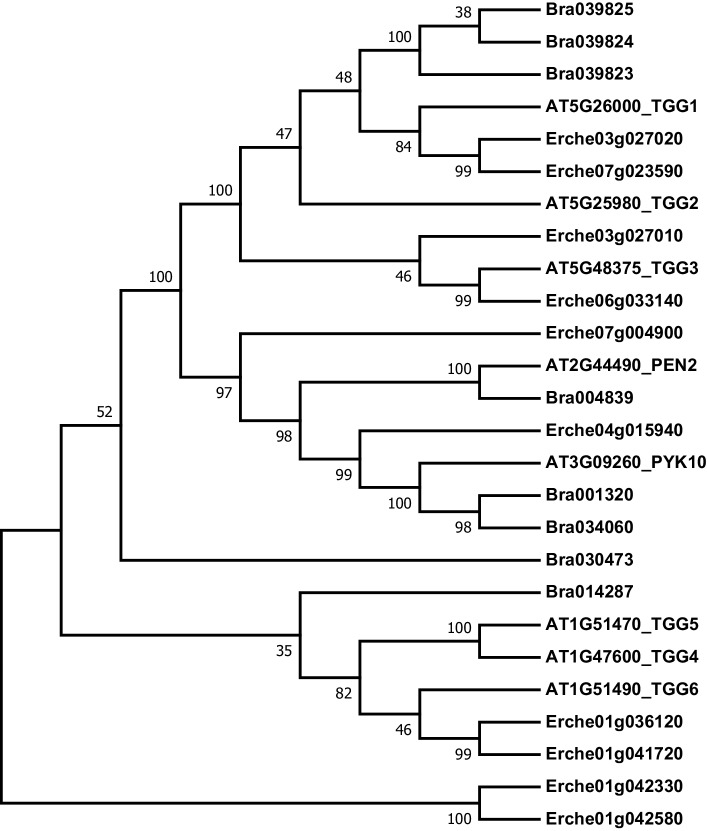
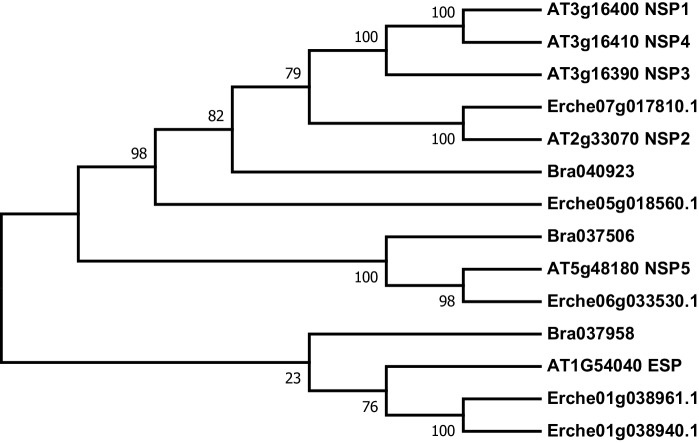
Figure 4. Identification of known aliphatic glucosinolate biosynthetic genes and glucosinolate-modifying genes from Arabidopsis in Erysimum cheiranthoides.
Aliphatic glucosinolates are synthesized from methionine by a series of enzymes, while additional enzymes are responsible for aliphatic glucosinolate modifications (black box). Red square brackets indicate where gene copy numbers differ between Arabidopsis and E. cheiranthoides, or where gene copies could not be matched unambiguously between species. Glucosinolates with names highlighted in blue were identified in Erysimum cheiranthoides var. Elbtalaue. Abbreviations: branched-chain aminotransferase (BCAT); bile acid transporter (BAT); methylthioalkylmalate synthase (MAM); isopropylmalate isomerase (IPMI); large subunit (LSU); small subunit (SSU); isopropylmalate dehydrogenase(IPMDH); cytochrome P450 monooxygenase (CYP); glutathione S-transferase F (GSTF); glutathione S-transferase Tau (GSTU); glutathione (GSH); γ-glutamyl peptidase 1 (GGP1); SUPERROOT 1 C-S lyase (SUR1); UDP-dependent glycosyltransferase (UGT); sulfotransferase (SOT); flavin monooxygenase (FMO); glucosinolate oxoglutarate-dependent dioxygenase (AOP); 3-butenyl glucosinolate 2-hydroxylase (GS-OH).