Table 1.
Structures and mechanism of action DNA-binding vanadium compounds (Kb-binding constant).
| Structure | Activity | References |
|---|---|---|
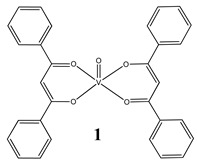
|
Groove binding to salmon sperm DNA accompanied with a partial insertion between the base stacks of the DNA (Kb = 2.3 × 103 M−1) Cytotoxicity (24 h): breast cancer cells MCF-7 (IC50 7.8 µM) liver cancer cells HepG2 (IC50 13.5 µM) colon cancer cells HT-29 (IC50 16.1 µM) |
[25] |
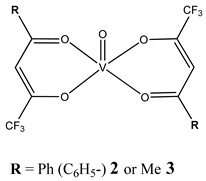
|
Oxidative cleavage of DNA through the generation of a hydroxyl radical Minor groove binding to DNA (2: Kb = 1.95 ± 0.16 × 103 M−1 3: Kb = 1.064 ± 0.17 × 103 M−1) Cytotoxicity (48 h): cervical cancer cells HeLa (2: IC50 256.9 µM 3: IC50 480.5 µM) |
[26] |
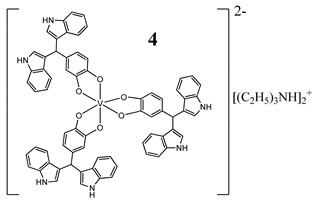
|
Similarities to cisplatin concerning DNA interaction ROS generation, mitochondrial damage, G2/M cell cycle arrest Cytotoxicity (72 h): panel of melanoma, colon, cervical, breast and pancreatic cancer cells IC50 < 10 µM for all cell lines |
[27] |
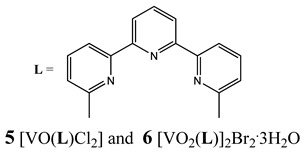
|
Intercalation as the way of DNA binding G2/M cell cycle arrest Cytotoxicity (48 h): cervical cancer cells HeLa (5: IC50 42.9 ± 1.5 µM 6: IC50 33.2 ± 0.9 µM) breast cancer cells T-47D (5: IC50 38.0 ± 1.6 µM 6: IC50 42.3 ± 1.8 µM) Lung cancer cells A549 (5: IC50 87.6 ± 2.4 µM 6: IC50 > 100 µM) |
[28] |
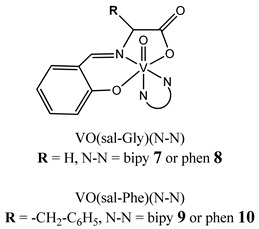
|
Phen-containing VIVO compounds display stronger DNA interaction ability than the corresponding bipy analogues Cytotoxicity (72 h): ovarian cancer cells A2780 (7: IC50 20.8 ± 0.5 µM 8: IC50 4.9 ± 1.3 µM 9: IC50 17.1 ± 3.9 µM 10: IC50 4.7 ± 1.8 µM) breast cancer cells MCF-7 (7: IC50 53 ± 2.0 µM 8: IC50 77 ± 1.3 µM 9: IC50 95 ± 3.7 µM 10: IC50 68 ± 1.4 µM) |
[29] |
|
Interaction with CT-DNA through a non-classical intercalative mode cleavage plasmid pBR322 DNA upon exposure to ultraviolet light Cytotoxicity (48 h): panel of cervical, breast and esophageal cancer cells IC50 range: 0.31–6.15 μM |
[30] |
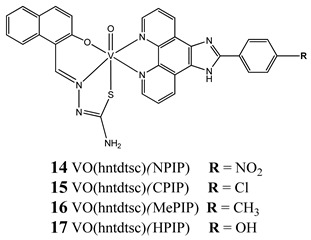
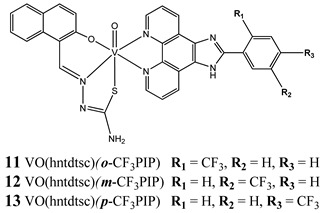
|
Binding with CT-DNA by an intercalation Kb = 14: 1.53 × 105 M−1 15: 1.41 × 105 M−1 16: 1.05 × 105 M−1 17: 0.95 × 105 M−1 cleave supercoiled plasmid DNA in the presence of H2O2 G0/G1 cell cycle arrest (14) Induction apoptosis in Hela cells (14) Cytotoxicity (24 h): cervical cancer cells HeLa (14: IC50 1.09 ± 0.16 µM 15: IC50 10.36 ± 1.23 µM) bladder cancer cell BIU-87 (14: IC50 4.51 ± 0.68 µM 15: IC50 8.69 ± 1.05 µM) lung cancer cells SPC-A-1 (14: IC50 7.61 ± 0.55 µM 15: IC50 21.43 ± 3.24 µM) |
[31] |
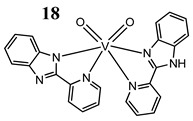
|
Interaction with DNA in a intercalative fashion (Kb = 2.76 × 105 M−1) Cytotoxicity (24 h): lung cancer cell A549 breast cancer cells MCF-7 keratinocyte cancer cell A431 IC50 for all cancer cell lines 75 μM normal human keratinocyte cells HaCaT IC50 150 µM |
[32] |
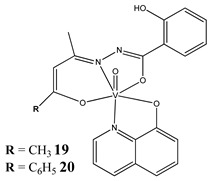
|
The intercalative mode of binding to DNA (19: Kb = 6.13 × 105 M−1 20: Kb = 8.69 × 105 M−1) Cytotoxicity (24 h): cervical cancer cell SiHa (19: IC50 33 µM 20: IC50 29 µM) |
[33] |
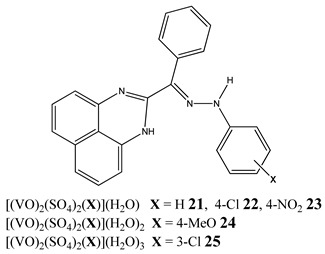
|
Binding to CT-DNA Kb = 21: 6.10 × 104 M−1 22: 7.99 × 104 M−1 23: 6.75 × 104 M−1 24: 6.07 × 104 M−1 25: 8.80 × 104 M−1 Cytotoxicity (48 h): breast cancer cells MCF-7 (25: IC50 11.44 µM 23: IC50 15.50 µM) liver cancer cells HepG2 (25: IC50 9.91 µM 23: IC50 11.01 µM) colon cancer cells HCT 116 (24: IC50 13.27 µM 23: IC50 15.53 µM) |
[34] |
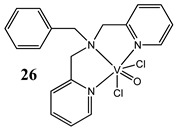
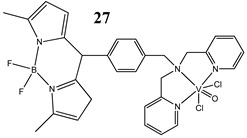
|
Light-activated VO2+-DNA crosslink formation (27) singlet oxygen (1O2) induced mitochondria-targeted PDT (27) Cytotoxicity (24 h): breast cancer cells MCF-7 (27: IC50 3.4±0.4 µM in visible light IC50 > 50 µM in the dark) cervical cancer cells HeLa (27: IC50 1.8±0.6 µM in visible light IC50 > 50 µM in the dark) 26: any significant cytotoxicity in light |
[36] |
|
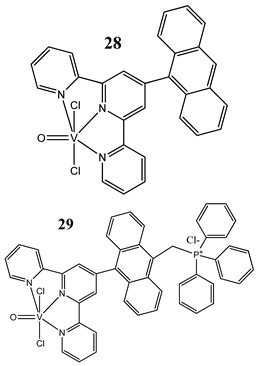
Light-activated DNA crosslink formation (in the dark they are partial DNA intercalators) ROS generation in visible light Cytotoxicity (24 h): breast cancer cells MCF-7 (28: IC50 10.4 ± 1.6 µM in visible light IC50 > 50 µM in the dark) (29: IC50 2.3 ± 0.3 µM in visible light IC50 27.6 ± 1.4 µM in the dark) cervical cancer cells HeLa (28: IC50 8.2 ± 0.3 µM in visible light IC50 > 50 µM in the dark) (29: IC50 1.8 ± 0.5 µM in visible light IC50 20.3 ± 1.0 µM in the dark) |
[37] |
|
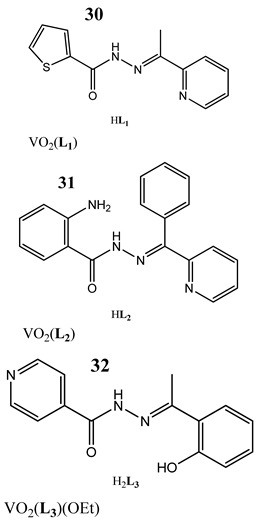
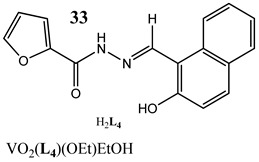
Photo-induced cleavage of pUC19 supercoiled plasmid DNA Interaction with CT-DNA through minor groove binding mode Kb = 30: 8.56 × 104 M−1 31: 1.13 × 105 M−1 32: 4.95 × 104 M−1 33: 5.03 × 103 M−1 Cytotoxicity (72 h): cervical cancer cells HeLa 30: IC50 20 ± 4.52 µM 31: IC50 18 ± 3.38 µM 32: IC50 19.5 ± 3.54 µM 33: IC50 9.9 ± 3.18 µM |
[38] |
|
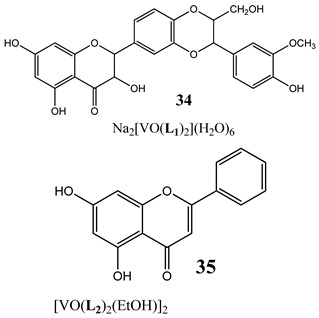
The topoisomerase IB inhibition (34) G2/M cell cycle arrest (35) activation caspase 3 and triggering the apoptosis (34) Cytotoxicity (24 h): colon cancer cells HT-29 A concentration-related inhibition from 75 to 100 µM |
[40] |
Bold and Underline: makes Table more readable.
