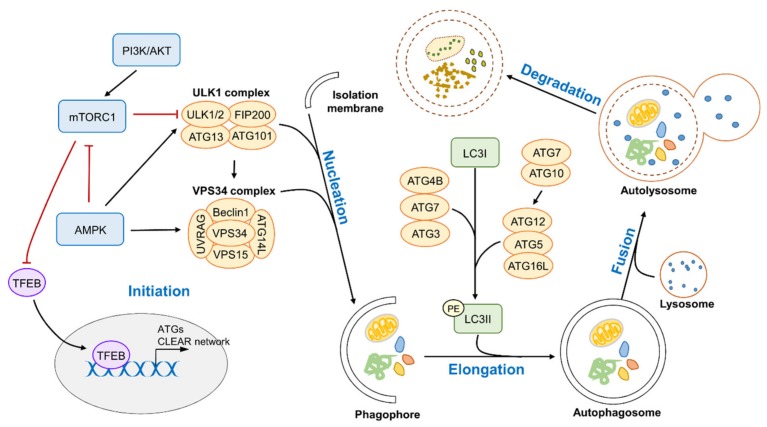
Figure 1.
The main pathway for autophagy modulation. The several groups of autophagy-related genes including ULK1 complex, VPS34 complex, and ATG12-ATG5-ATG16L complex involved in autophagosome formation. Depletion of growth factor and/or nutrient cause the inactivation of mammalian target of rapamycin complex 1 (mTORC1) that acts as an autophagy suppressor by inhibiting ULK1 genes. AMP-activated protein kinase (AMPK) positively modulates autophagy machinery via inhibition of mTORC1 and activation of either ULK1 complex or VPS34 complex. ER-stress mediated unfolded protein response (UPR) stimulates AMPK-driven autophagy and is also involved in the process for autophagosome formation (not shown). Activated ULK1 complex resulted in activating VPS34 complex, facilitating the autophagy process including nucleation and elongation. Conversion of LC3I into phosphatidylethanolamine (PE)-LC3II, a key substrate for autophagosome membrane assembly, is mediated by two ubiquitin-like conjugation systems including ATG4B, ATG7, ATG3, and ATG12-ATG5-ATG16L complex. Subsequently, closed autophagosome fuses with lysosome to form an autolysosome, which leads to autophagy degradation of cytoplasmic components. This process is sustained by preventing the mTORC1-mediated phosphorylation of transcription factor EB (TFEB), which induces the expression of genes involved in autophagosome and lysosome biogenesis. CLEAR, coordinated lysosomal expression and regulation motif; PI3K, phosphoinositide-3-kinase; AKT, protein kinase B; UVRAG, UV radiation resistance-associated gene protein.