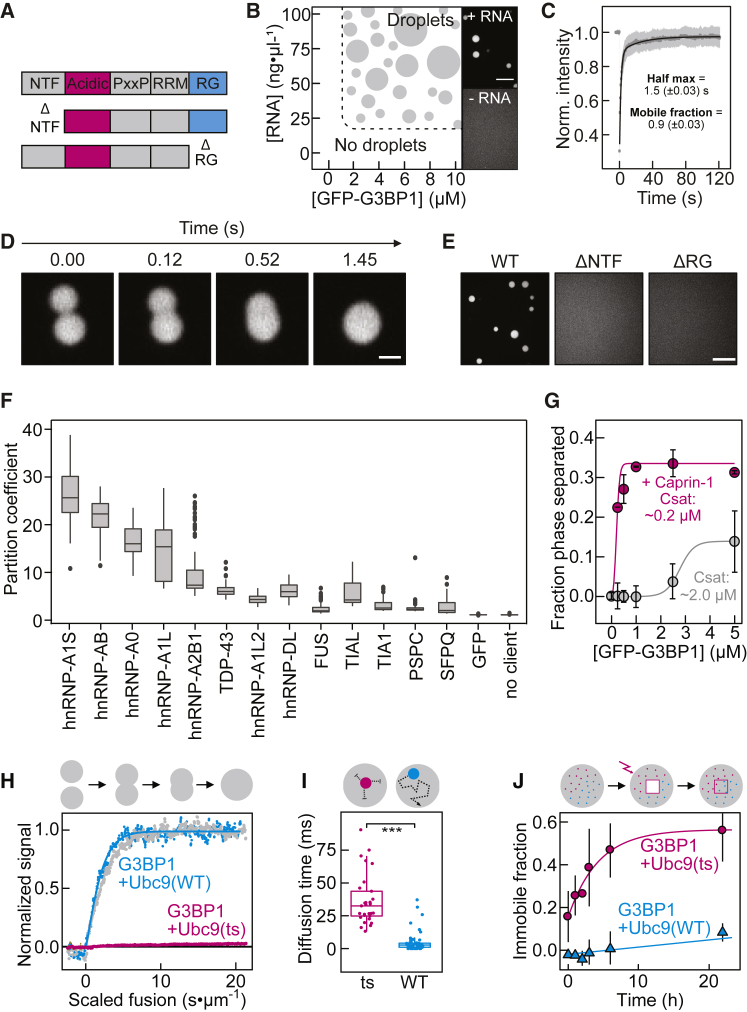
Figure 2.
In Vitro Reconstituted G3BP1 Condensates Recapitulate Cellular SG Properties
(A) Schematic domain structure of G3BP1.
(B) Phase diagram of G3BP1(WT) as a function of protein and RNA concentration. Right: fluorescence images of G3BP1(WT) with and without RNA.
(C) Analysis of in vitro partial FRAP of G3BP1(WT)-RNA condensates. Mean average data (gray dots), fit (black), SD (light gray), n = 20.
(D) Fluorescence images from a time-lapse video of G3BP1(WT)-RNA condensate fusion.
(E) Fluorescence images of G3BP1 variants with RNA.
(F) Partition coefficient of GFP-tagged RBPs in preformed SNAP (Alexa 546)-labeled G3BP1-RNA condensates. PSPC, SFPQ, and GFP served as negative controls (n = 150 fields of view [FOVs]) (Figure S2H).
(G) G3BP1(WT) saturation concentration (Csat) with and without mCherry-Caprin-1 (mean, SD, fit, n = 20 FOVs).
(H) Coalescence of G3BP1(WT)-RNA condensates with or without equimolar Cy3-labeled Ubc9(WT) or Ubc9(ts), measured with dual-trap optical tweezers.
(I) Diffusion time of Ubc9(WT) and Ubc9(ts) within G3BP1-RNA condensates, determined by FCS. Significance levels: ∗ < 0.05, ∗∗ < 0.01, ∗∗∗ < 0.001.
(J) Mean average immobile fraction of Ubc9(WT) and Ubc9(ts) within G3BP1-RNA condensates as a function of time, determined by FRAP (SD, n = 5). Condensates formed with 1% PEG-20K and, when specified, 75 ng/μL of total RNA.
Scale bars, 10 μm, except 1 μm in (D).