Figure S3.
Related to Figure 3
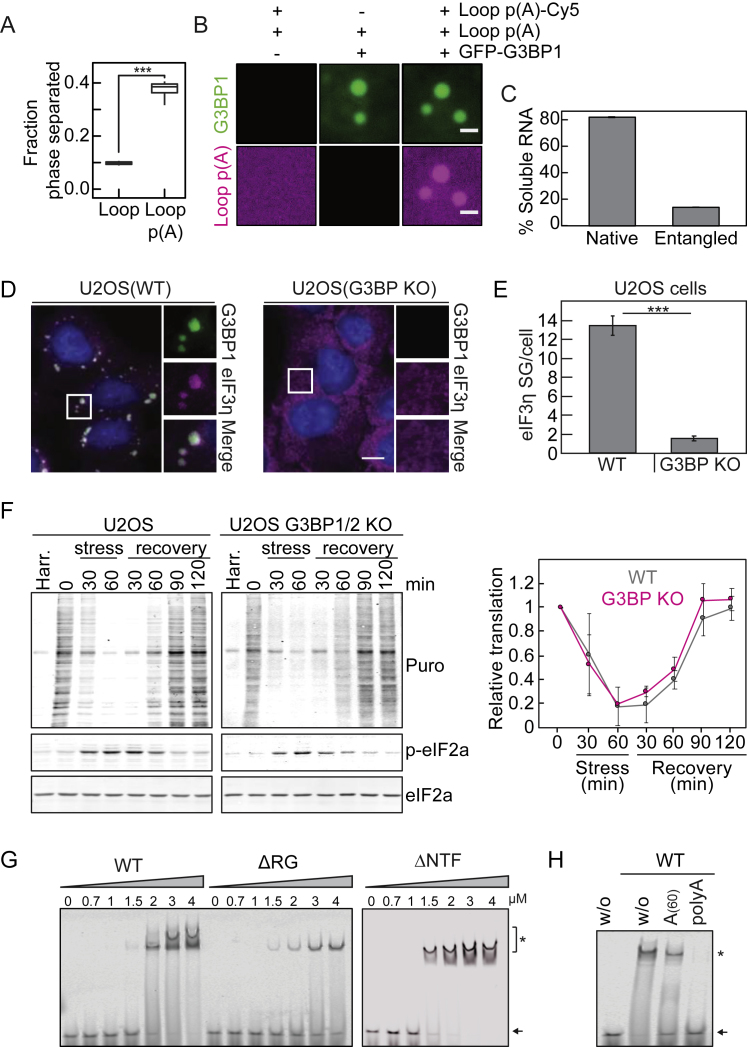
A: Phase separated fraction of G3BP1(WT) in the presence of 5-Loop and polyadenylated 5-Loop RNAs (n = 8 FOV). B: Fluorescence images of GFP-G3BP1(WT)-RNA condensates formed with polyadenylated and Cy5-labeled 5-Loop RNA. Scale bar, 5 μm. Condensates in A-B were formed in the presence of 1% PEG-20K and 75 ng/μl RNA. C: Quantification of the soluble fraction of RNA remaining after tangle formation (mean and SD from three independent measurements). D: Fluorescence immunostaining of oxidatively stressed U2OS wild-type cells and U2OS G3BP1/2 KO cells. G3BP1 (green) and eIF3η (magenta) are shown. Scale bars, 10 μm. E: Quantification of the number of eIF3η-positive SGs per cell in U2OS wild-type and in U2OS G3BP1/2 KO cells (mean and SD, n = 184 - 259 cells). F: Translation levels in U2OS cells (WT or G3BP1/2 KO) upon oxidative stress and recovery from stress. Translation levels were assessed by puromycin incorporation. Immunoblots against puromycin, p-eIF2a (to follow stress kinetics) and eIF2a (loading control) are shown. Harringtonine (Harr.) was used to prove specific incorporation of puromycin into nascent polypeptide chains. On the right, the normalized translation levels upon stress and recovery are shown (mean and SD from three independent experiments). G: EMSA to determine the apparent binding affinity of G3BP1(WT), G3BP1(ΔRG) and G3BP1(ΔNTF2) to A(60)-Cy5 RNA. Black arrow points toward free A(60)-Cy5 RNA. ∗ indicates shifted RNA species due to G3BP1 binding. H: EMSA testing for the competition of unlabeled A(60) and poly(A) (500-4000 nt) RNAs for G3BP1(WT) binding to the A(60)-Cy5 probe. From left to right: Lane1: without G3BP1, Lane 2: without competitor, Lane 3: with unlabeled A(60) RNA as competitor, Lane 4: with unlabeled poly(A) as competitor. Black arrow points toward free A(60)-Cy5 RNA. ∗ indicates shifted RNA species due to G3BP1 binding.