Figure S5.
Related to Figure 5
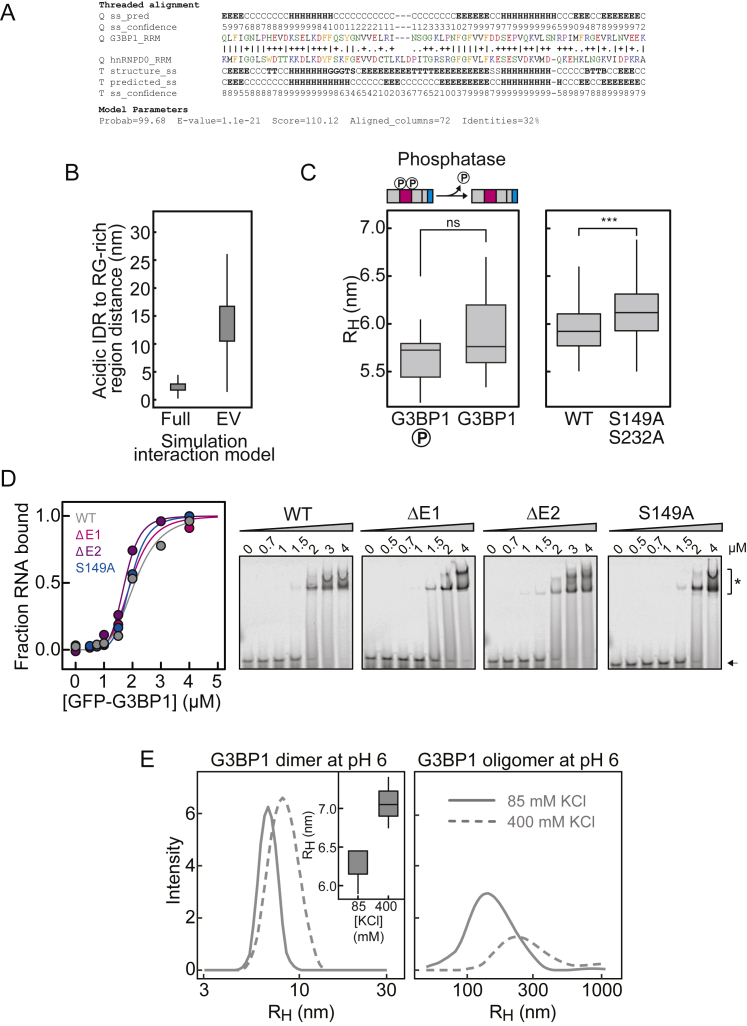
A: Homology model sequence alignment for G3BP1-RRM. Model parameters are robust, and the resulting homology model is structurally reasonable and high-confidence. B: The intramolecular distances between the acidic tract and the RG-rich region are substantially lower in full Hamiltonian simulations (Full) than would be expected based on a generic self-avoiding description of the IDR (Excluded Volume, EV). This result highlights the importance of intramolecular attractions between the acidic tract and the RG-rich region. C: Left, comparison of the hydrodynamic radius (RH) of phosphorylated G3BP1(WT) (indicated with ‘P’) and dephosphorylated G3BP1(WT). Right, RH of phosphorylated G3BP1(WT) and G3BP1(S149A/S232A). RH was determined by FCS. D: Right, EMSA to determine the apparent binding affinity of G3BP1(WT), G3BP1(ΔE1), G3BP1(ΔE2) and G3BP1(S149A) to A(60)-Cy5 RNA. Black arrow points toward free A(60)-Cy5 RNA. ∗ indicates shifted RNA species due to G3BP1 binding. The quantification of the EMSAs is shown on the left (G3BP1(WT): KD ~2.0 μM; G3BP1(S149A): KD ~1.9 μM; G3BP1(ΔE1): KD ~1.9 μM; G3BP1(ΔE2): KD ~1.7 μM). E: RH of G3BP1(WT) at pH 6 and 85 mM KCl and after increasing the KCl concentration to 400 mM, determined by DLS (mean average of 6 measurements). On the left, the increase of RH of the G3BP1(WT) dimer upon increasing KCl concentration is shown. On the right, the oligomeric species of G3BP1(WT) are depicted. Note that the intensity of the oligomer decreases at 400 mM KCl.