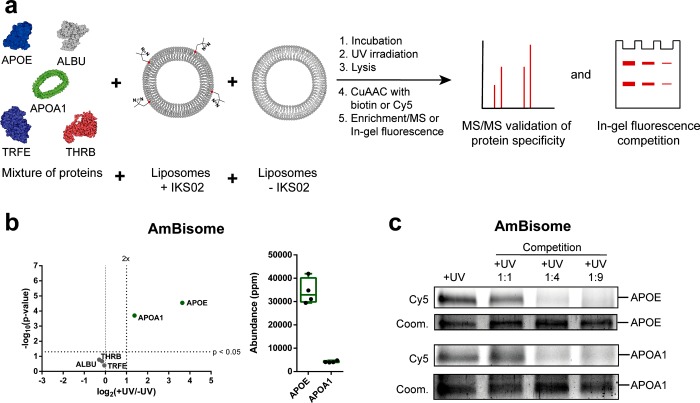
Figure 4.
Validation of apolipoprotein E and A1 binding to AmBisome liposomes. (a) Liposomes were incubated in a mixture of purified human serum proteins consisting of apolipoprotein E (APOE, 2 μgmL–1), serum albumin (ALBU, 25 μgmL–1), apolipoprotein A-I (APOA1, 2 μgmL–1), transferrin (TRFE, 10 μgmL–1), and prothrombin (THRB, 2 μgmL–1). (b) Volcano plot of protein enrichment over background (log2(+UV/–UV)) plotted against the statistical significance of this comparison (−log10(p-value)). Proteins meeting all selection criteria labeled in green. Abundance plot displaying the abundancies of apoE and apoA1 within the +UV samples. (c) Competition assay of apolipoprotein E and A1 binding. Increasing concentrations (1:1 to 1:9 molar ratios) of unlabeled AmBisome liposomes were incubated, together with AmBisome liposomes, containing IKS02, in the above predefined mixture of human serum proteins. Captured apoE and apoA1 on the surface of IKS02-labeled AmBisome liposomes were separated by SDS-PAGE and visualized by in-gel fluorescence (Cy5). Protein loading determined by Coomassie Blue (coom.). Protein structures were obtained from the protein data bank (PDB): (APOE: 2L7B, APOA1: 1AV1, ALBU: 1E78, THRB: 6C2W, TRFE: 1D3K). Illustrations were generated using Illustrate.48