Fig. 1 |. Identity and sequence contexts of rNMP incorporation in S. cerevisiae genome.
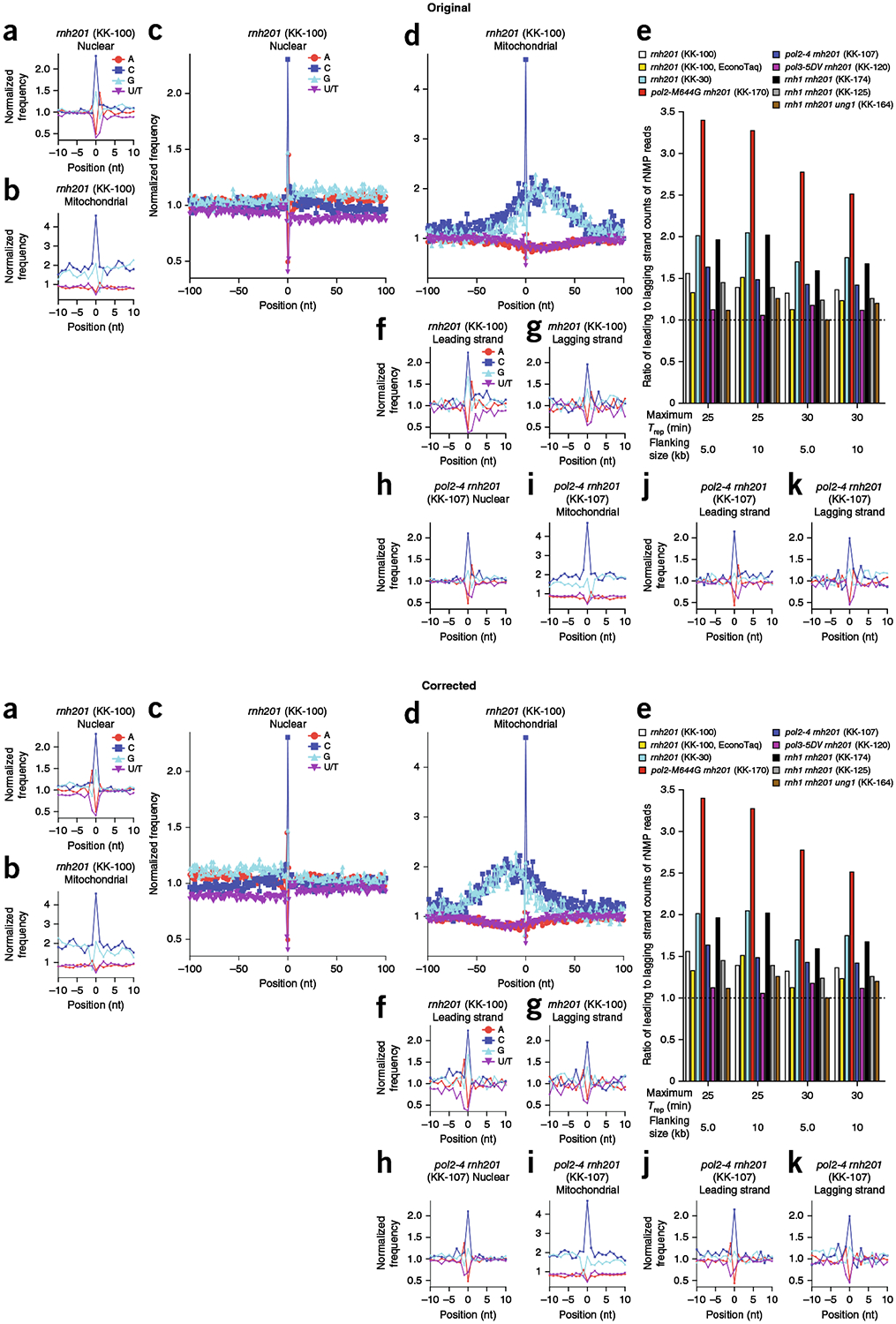
a,b, Normalized nucleotide frequencies relative to mapped positions of sequences from Ribose-seq library of rnh201Δ (KK-100) cells. Position 0 is the rNMP. c,d, Zoom-out of frequencies. e, Ratios of rNMPs on newly synthesized leading to lagging strand for all ribose-seq libraries. Early-firing ARSs selected by their replication timing (Trep) were investigated for two different flanking sizes. f,g, Normalized nucleotide frequencies relative to mapped sequence positions in leading (f) and lagging (g) strands from the rnh201Δ (KK-100) library. ARSs with a Trep of no longer than 25 min were selected with flanking size of 10 kb. h–k, Normalized nucleotide frequencies relative to mapped sequence positions from a pol2-4 rnh201Δ (KK-107) library. Reads were mapped to (a,c,h) nuclear genome, (b,d,i) mitochondrial genome, (f,j) leading strand or (g,k) lagging strand.
