Figure 1. Schematic of the YAC breakage assay.
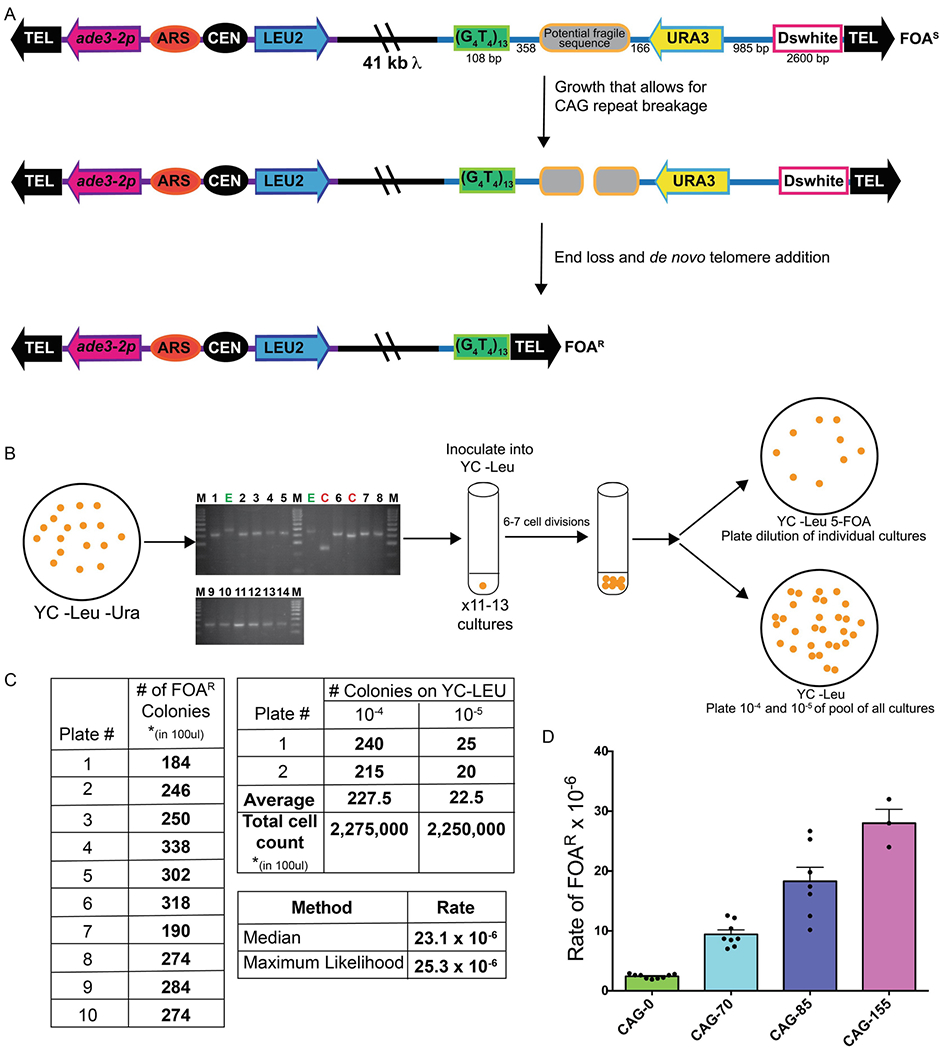
A. In this system, a potential fragile sequence (such as an expanded CAG repeat tract) is integrated onto a yeast artificial chromosome between a telomere seed sequence (G4T4)13 and the URA3 gene. Breaks that occur within the fragile sequence are subject to resection and telomere addition, which results in loss of the URA3 gene and renders cells 5-FOAR. The YAC additionally contains a LEU2 marker gene, which allows for maintenance of the YAC, a centromere (CEN4) and an origin of replication (ARS1). The blue line indicates the pYIP5 plasmid backbone, the black line indicates lambda DNA, and the purple line corresponds to pUC18 plasmid backbone. B. In the YAC end loss assay, strains are plated for single colonies on YC –Leu –Ura plates and 10 individual colonies of the correct tract length (if testing an unstable repeat sequence) are grown in YC –Leu for 6-7 cell divisions. A portion of each individual culture is plated onto YC –Leu +5-FOA plates, and a portion of the culture is pooled and serially diluted to obtain single colonies on YC –Leu, which serves as the total viable cell count. C. Example of the calculation of fragility rate for a 10-colony YAC CF1 CAG-85 fragility assay. Colony counts are used to determine the number of mutants and the number of viable cells in 100 μl of logarithmically growing cultures. D. Wild-type fragility data of S. cereviase strain BY4705 where the fragile sequence integrated between the G4T4 and URA3 marker are different tract lengths of CAG repeats. Assays done with this (CAG)n-URA3 YAC show that 5-FOA resistance increases with increasing number of CAG repeats. Data sourced from (8, 27–30).
