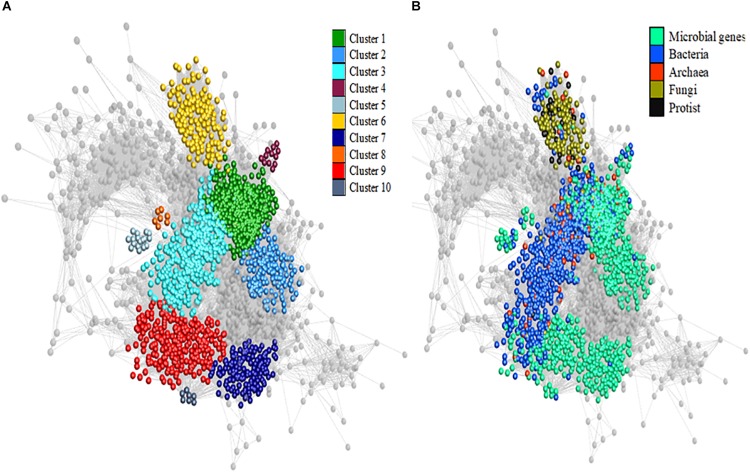
FIGURE 1.
Functional clusters composed of microbial genera and genes generated using co-abundance network analysis in beef cattle. (A) Distribution of the clusters in the network. (B) Distribution of genes and microbial genera (bacteria, archaea, fungi, and protist) among the clusters. Nodes represent microbial genera and genes, and edges illustrate co-abundances between their relative abundances. Networks were clustered using the MCL algorithm, and clusters 1 to 10 are shown. Only variables with correlation values greater than 0.70 between nodes were kept during the analysis. Cluster 1 containing most abundant methanogenic archaea (Methanobrevibacter, Methanosphaera, and Methanosarcina), and microbial genes involved in methanogenesis pathway, and also bacteria (Sarcina), fungi (Tremella) and genes in degradation pathways for amino acids (nitrogen fixation capacity of Candidatus Azobacteroides) and carbohydrates, was referred to as methanogenesis cluster. Cluster 2 includes only genus Fibrobacter and microbial genes involved in the synthesis of central metabolic enzymes. Cluster 3 is mainly comprised of bacteria of the phyla Firmicutes, Proteobacteria, and Acidobacteria with low abundant archaea, some of them methanogen. Cluster 4 is a small cluster containing Butyrivibrio, Pseudobutyrivibrio and few microbial genes related to sugar metabolism. Cluster 5 is also a reduced cluster containing Bacillus, other bacteria and genes related to sugar degradation. Cluster 6 is dominated by genera of the fungal community, and three hydrogenotrophic and/or acetoclastic methanogens. Cluster 7 included Bifidobacterium and microbial genes relevant for carbohydrate degradation. Cluster 8 contained Prevotella with genes involved in nitrogen metabolism and pentose phosphate pathway. Cluster 9 contained the methylotrophic Methanomassiliicoccales Candidatus Methanomethylophilus, the acetogens Eubacterium, Blautia, and Acetitomaculum and a high diversity of Proteobacteria (mainly γ-Proteobacteria) and microbial genes involved in carbohydrates, lipids, and aminoacids metabolism. Cluster 10 includes Selenomonas and few microbial genes related to oligosaccharide transport.