Figure 1.
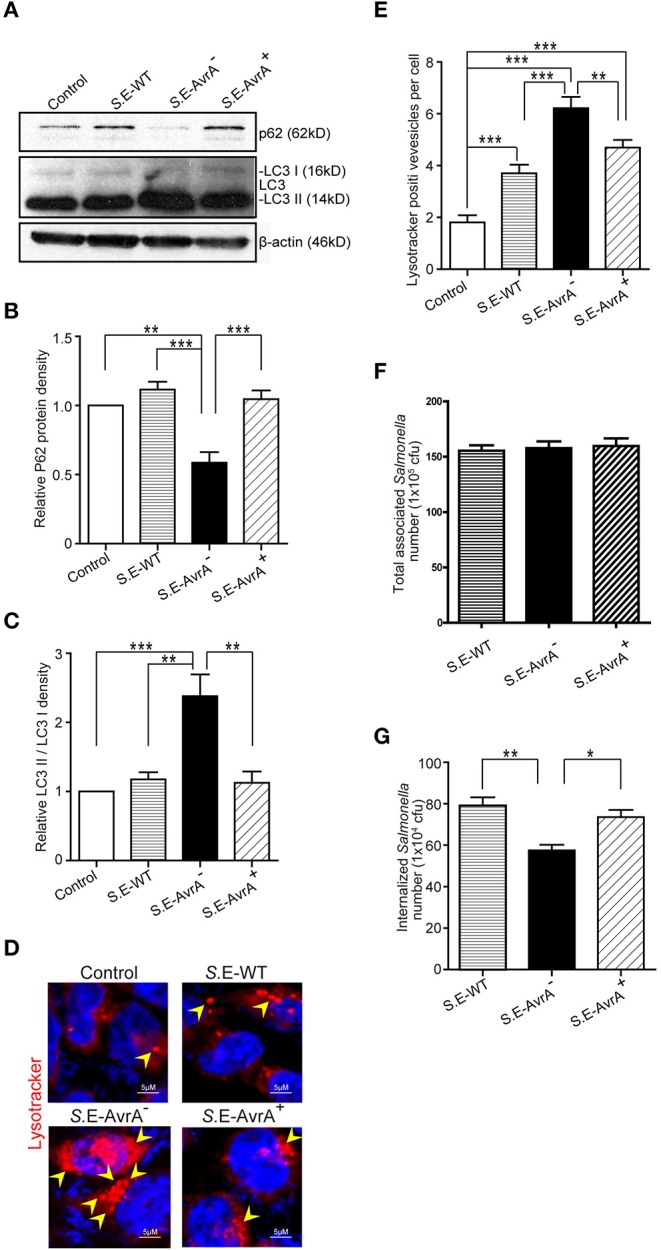
Salmonella Enteritidis AvrA inhibits autophagy in cell models. AvrA-regulated expression levels of autophagy related proteins. (A) The indicated HCT116 cell lines were infected with S.E WT, S.E-AvrA− and S.E-AvrA+ strains (MOI:1:1, 1 h incubation before harvested, n = 4) as shown and analyzed for protein expression by immunoblotting. The immunoblotting of P62/LC3 was used to track the expression of P62 and the conversion of LC3 I into LC3 II for autophagic activity in the HCT116 cells after infection with the different S.E. The relative density of P62 (B) and LC3 II/LC3 I (C) was determined using Quantity One 4.6.2 software (Bio-Rad, Hercules, CA, USA). N = 4, **adjusted P < 0.01, ***adjusted P < 0.001 by ANOVA test. (D) HCT116 cell lines were incubated with 100 nM LysoTracker Deep Red Probe, then infected with S.E WT, S.E-AvrA- and S.E-AvrA+ strains (MOI:1:1, 1 h incubation before microscopic examination) to check the lysosomes staining. The immunofluorescence indicated that the HCT116 cells, pre-treated with LysoTracker, showed more lysosomes in the cells infected with the S.E-AvrA− bacteria compared with the cells infected by the AvrA present strains. (E) Quantification of the number of lysotracker positive vesicles. The data are reported as the mean ± SE from three independent experiments, and a total of 100 cells per condition were analyzed. n = 100, **adjusted P < 0.01, ***adjusted P < 0.001 by ANOVA test. The numbers of associated Salmonella and internalized Salmonella in human epithelial cells colonized with wild-type Salmonella Enteritidis or AvrA mutant or AvrA-complemented strains. Human epithelial cells were grown on an insert, colonized with an equal number of the indicated bacteria for 30 min, after washed with HBSS, the number of associated Salmonella (Salmonella adhesion) was determined (F). Alternatively, cells were incubated in DMEM containing gentamicin (100 μg/ml) for 30 min, and the number of internalized intracellular Salmonella (Salmonella invasion) was then determined (G). Data are reported as the mean ± SE from six independent experiments, *adjusted P < 0.05, **adjusted P < 0.05 by ANOVA test.
