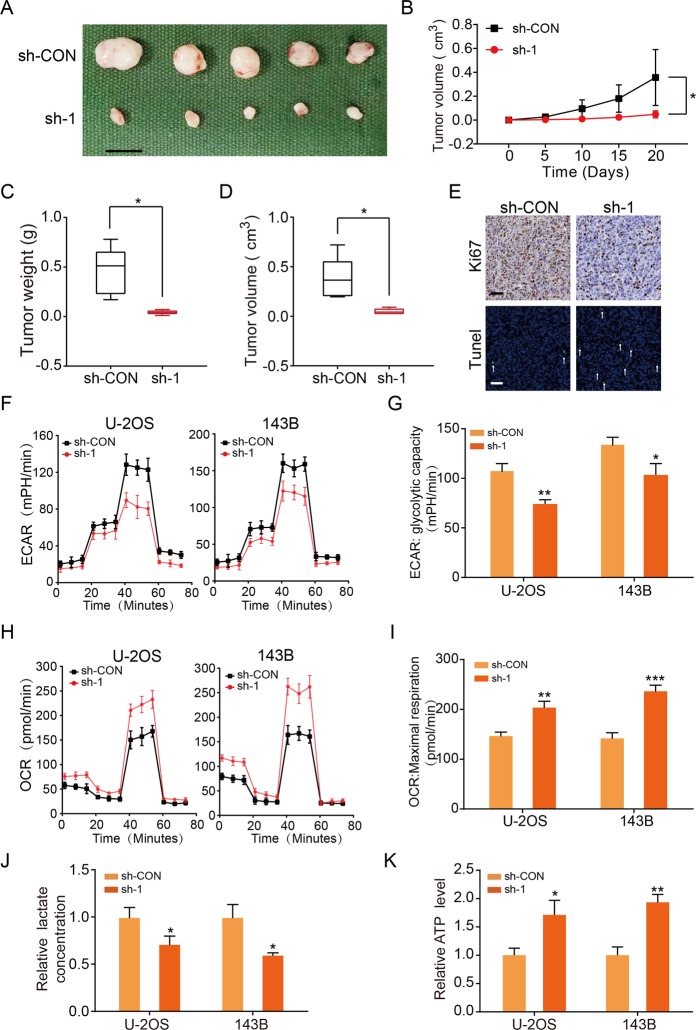
Fig. 2. KCNQ1OT1 facilitates OS proliferation in vivo and promoted aerobic glycolysis in OS cells.
a Morphologic characteristics of xenograft tumors from U-2OS/sh-Control group and U-2OS/sh-KCNQ1OT1 group (n = 5). Scale bars = 1 cm. b The tumor volumes were measured with calipers every 5 days. Values are means ± SD, *p < 0.05 (Student’s t-test). c Tumor weights at 20 days were measured in each group. The median, upper, and lower quartiles were plotted, and the whiskers that extend from each box indicate the range of values that were outside of the intra-quartile range. n = 5, *p < 0.05 (Student’s t-test). d Tumor volumes at 20 days were measured in each group. The median, upper, and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range. n = 5, *p < 0.05 (Student’s t-test). e Representative images of Ki67 and TUNEL staining in the xenograft tumors from the sh-KCNQ1OT1 and sh-Control mice. A TUNEL positive cell is indicated (arrow). f Extracellular acidification rate (ECAR) of U-2OS or 143B cells in sh-Control and the sh-KCNQ1OT1 group was detected via a Seahorse Bioscience XFp analyzer. Glc: glucose, Oligo: oligomycin, 2-DG: 2-deoxy-d-glucose. g Quantification of the glycolytic capacity from the Fig. 2f, *p < 0.05, **p < 0.01 (Student’s t-test). h O2 consumption rate (OCR) of U-2OS or 143B cells in sh-Control and sh-KCNQ1OT1 group was detected using a Seahorse Bioscience XFp analyzer. O: Oligomycin, F: FCCP, A&R: antimycin A/rotenone. i Quantification of maximal respiration from the Fig. 2h, **p < 0.01, ***p < 0.001 (Student’s t-test). j Lactate production was determined in the U-2OS and 143B cells stable knockdown KCNQ1OT1. Values are means ± SD, *p < 0.05 (Student’s t-test). k ATP level was determined in U-2OS and 143B cells stable knockdown KCNQ1OT1. Values are means ± SD, *p < 0.05, **p < 0.01 (Student’s t-test).