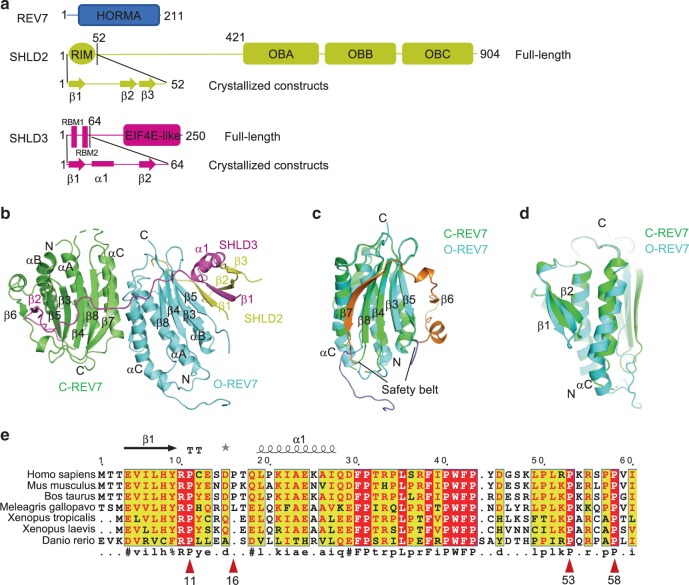
Fig. 1. Overall structure of the SHLD3-REV7-SHLD2 ternary complex.
a Schematic representation showing the organization of REV7, SHLD2 and SHLD3. The secondary structures of the truncations of SHLD2 and SHLD3 used for crystallization are shown in detail, arrow indicates β strand and rectangle represents α helix. HORMA, a domain named after the Hop1, REV7 and Mad2 proteins; RIM, REV7 interacting motif; RBM, REV7 binding motif, characterized as PXXXpP. b Structure of the SHLD3-C-REV7-O-REV7-SHLD2 complex. Two REV7 molecules are differentially colored to indicate their different states, C-REV7 is shown in green and O-REV7 is shown in cyan. The secondary structures of C-REV7, O-REV7, SHLD2 and SHLD3 are labeled. Disordered loop is shown as dashed lines. c Structural alignment of C-REV7 and O-REV7. The regions between two lines represent the safety belt and are colored in orange (C-REV7) and slate (O-REV7). d Structural alignment of C-REV7 and O-REV7 viewed in another side, which shows the different positions of αC, β1, β2 between C-REV7 and O-REV7. e Sequence alignment of SHLD3(1–60) across species. The highly conserved residues are shown in red background. The prolines which are proposed as the conserved PXXXpP motif are indicated with red triangle and numbered.