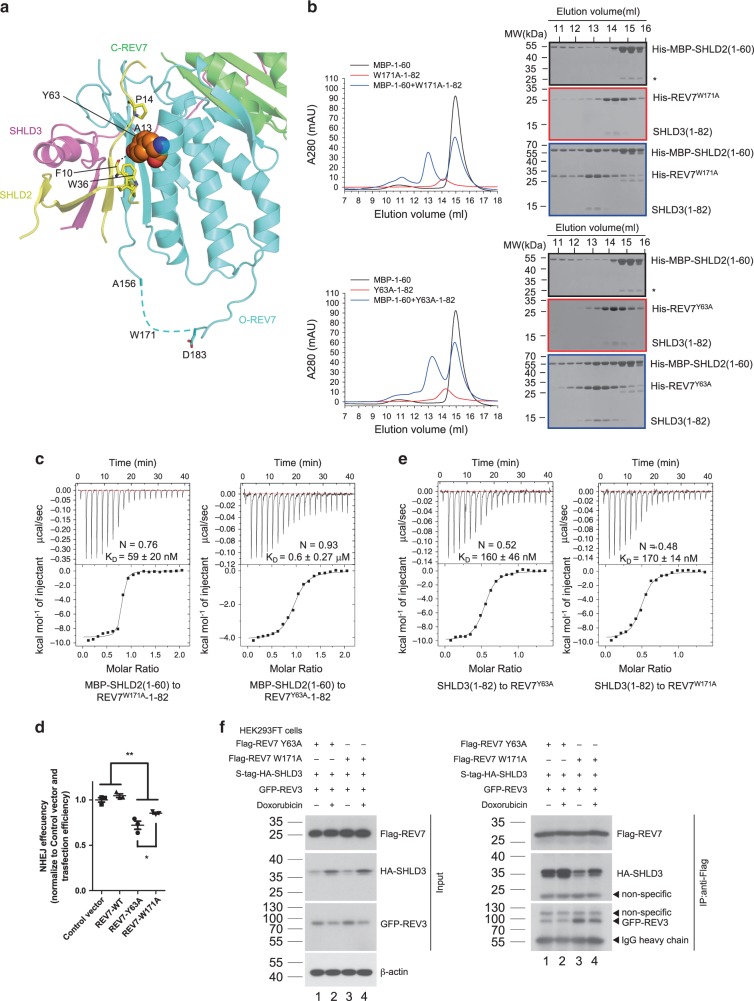
Fig. 7. REV7Y63A shows impaired ability to interact with SHLD2 but not SHLD3.
a Structural details of the interface between O-REV7 and SHLD2. C-REV7 is shown in green, O-REV7 is shown in cyan, SHLD3 is shown in magenta and SHLD2 is shown in yellow. Tyr63 of O-REV7 are shown in spheres model to highlight its interaction with SHLD2. The residues Phe10, Ala13, Pro14 and Trp36 of SHLD2 that make interactions with Tyr63 are shown in sticks. Trp171 of O-REV7 locates in the disordered region shown as dashed lines. b Gel filtration profiles show the interaction between MBP-SHLD2(1–60) (short as MBP-1–60) and REV7Y63A-SHLD3(1–82)/REV7W171A-SHLD3(1–82) (short as Y63A-1–82/W171A-1–82) in a Superdex200 Increase 10/300 SEC column. MBP-SHLD2(1–60) is excessive. Abs 0.1% of REV7Y63A-SHLD3(1–82), REV7W171A-SHLD3(1–82), and MBP-SHLD2(1–60) are 0.534, 0.405, and 1.668, respectively. This indicates extinction coefficient of REV7-SHLD3(1–82) is low, which accounts for its lower A280. Asterisk represents impurity. (n = 2). c ITC measurements of interaction between REV7Y63A-SHLD3(1–82) or REV7W171A-SHLD3(1–82) and MBP-SHLD2(1–60). The calculated N and KD are indicated as described in Fig. 2d. d NHEJ efficiency was determined in Hela cells overexpressing exogenous FLAG-tagged wild-type REV7, REV7-Y63A, REV7-W171A or control vector. Data were analyzed with the unpaired two-tailed Student’s test. Error bars indicate SEM (n = 3). *p < 0.05, **p < 0.01. Exact p-values are 0.0056 (Control versus REV7-Y63A), 0.0054 (Control versus REV7-W171A), 0.0028 (REV7-WT versus REV7-Y63A), 0.0011 (REV7-WT versus REV7-W171A) and 0.045 (REV7-Y63A versus REV7-W171A). e ITC measurements of interaction between SHLD3(1–82) and REV7Y63A or REV7W171A. The calculated N and KD are indicated as described in Fig. 2d. f S-tag-HA-SHLD3 and Flag-REV7 mutants were co-expressed with GFP-tagged REV3(1042–1251 + 1847–2021) (short as GFP-REV3 in the Figure) in HEK293FT cells. Indicated cells were treated with DMSO, or 1 μM doxorubicin for 24 h, then lysed with NP-40 lysis buffer. Flag-REV7 and its associated proteins were purified by anti-Flag agaroses. HA, Flag and GFP antibodies were used to detect S-tag-HA-SHLD3, Flag-REV7 and GFP-tagged REV3(1042–1251 + 1847–2021), respectively. The input panel shows the transfection efficiency and the IP panel shows the interaction between REV7 mutants and SHLD3 or REV3(1042–1251 + 1847–2021). (n = 2). n, biologically independent experiments. Source data are provided as a Source Data file.