Figure 2.
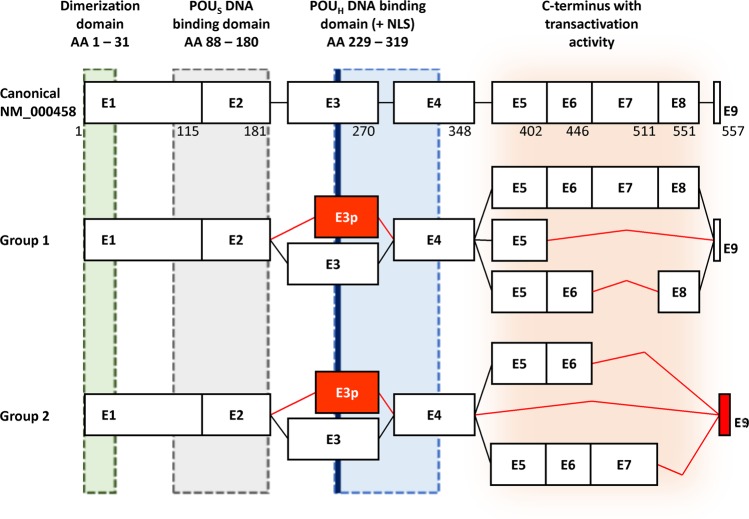
Scheme of the proposed HNF1B splicing pattern HNF1B alternative splicing variants can be divided into two main groups according the maintenance of original reading frame. ASVs in the first group maintain original reading frame across the whole transcript, including last exon 9, while ASVs in second group cause exon 9 frameshift (red box) which results in the use of alternative STOP codon (92 bp after original STOP codon). We propose, that all ASVs in each group will be present in both forms of exon 3, canonical or alternatively spliced (e3p; red box). Black lines connecting exon boxes represents canonical transcript, while red lines represent alternative splicing event. The lengths of the exons are proportional. The white boxes illustrate the unaffected coding exons. Corresponding amino acid (AA) numbers are indicated below the exon boxes of canonical isoform. The green, grey, blue and orange areas illustrate the coding areas for functional domains across the HNF1B transcripts. NLS – nuclear localization signal (thick blue line). POUS – POU specific domain. POUH – POU homeodomain. The scheme was adopted2 and modified.