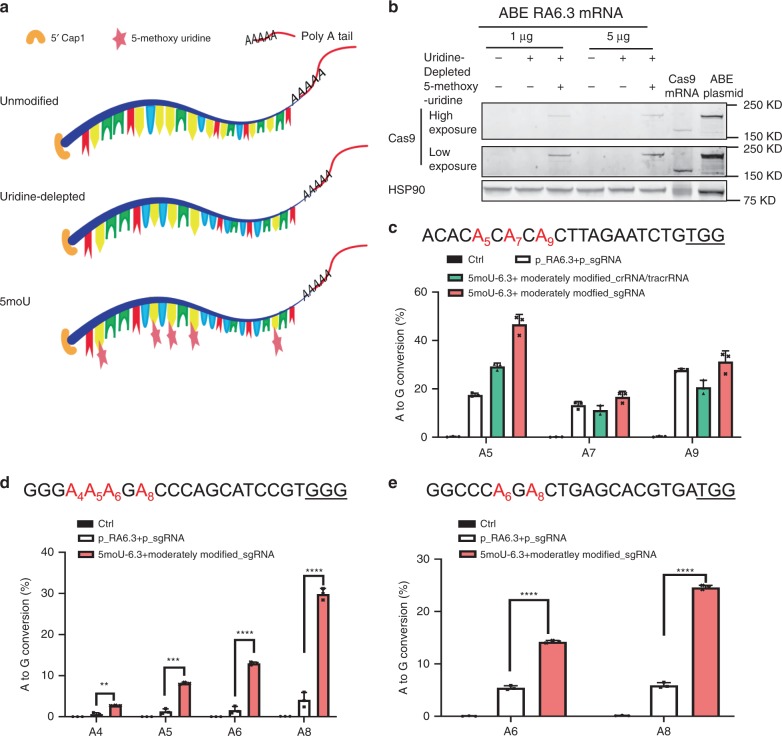
Fig. 1. Chemical modifications of ABE mRNA and guide RNAs are critical for efficient base editing in cells.
a Diagrams of ABE RA6.3 mRNAs with sequence-optimization (uridine-depletion) and chemical modification (5-methoxyuridine). Red: “A”; Yellow: “U”; Green: “G”; Blue: “C”. b ABE mRNA with both 5-methoxyuridine modification and uridine-depletion showed the highest protein expression in HEK293T cells by western blot. The mass of transfected RA6.3 mRNA is as indicated. ABE plasmid and Cas9 mRNA served as controls. c Comparison of editing efficiency by plasmids of ABE and guide RNA (p_RA6.3 + p_sgRNA), ABE mRNA + tracrRNA/crRNA, and ABE mRNA + sgRNA in HEK293T cells. The target “A” sites are highlighted in red. PAM is underlined. Data represent mean ± SD. d, e, Comparison of editing efficiency by DNA and RNA-encoded ABE at two genomic sites in HEK293T cells. The target “A” sites are highlighted in red. PAM is underlined. c–e All mRNAs are 5moU-6.3. All guide RNAs are moderately modified RNA. Control group (Ctrl) is cells transfected with 500 ng GFP plasmid. Graphs show mean values. Data represent mean ± SD (n = 3 biologically independent samples). d P = 0.0079 (**), 0.0005(***), <0.0001 (****), <0.0001 (****); e P < 0.0001 (****) (two tailed t-test). Source data are provided as a Source Data file for b–e.