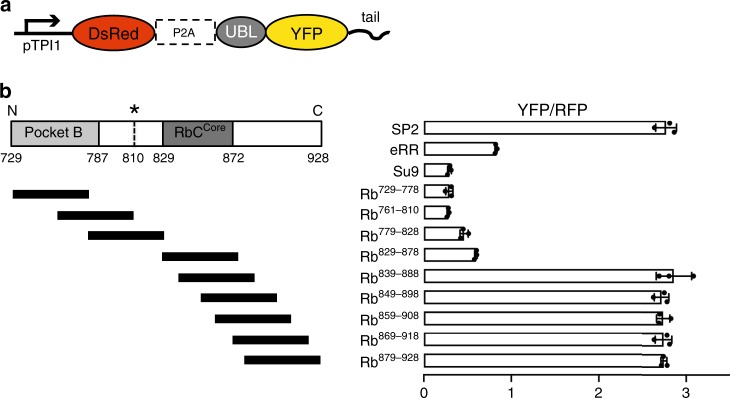
Fig. 3. Proteasome initiation at different Rb sequences.
a Schematic representation of the fluorescence-based degradation assay in S. cerevisiae. The substrate proteins consisted of an N-terminal UBL domain derived from S. cerevisiae Rad23, followed by a YFP domain and a tail. Genes encoding DsRed (RFP) followed by the ribosome skipping sequence P2A and the coding sequence for YFP were expressed from the same promoter (pTPI1) on the same plasmid to produce the RFP reference and YFP substrates at a constant ratio. b Cell fluorescence profiles of S. cerevisiae cultures expressing proteasome substrates with different initiation regions monitored by flow cytometry. UBL-YFP substrates with control initiation regions (SP2, eRR, and Su9) or 50 amino acid sequences derived from Rb as indicated. The graph shows standardized median cellular YFP fluorescence (median YFP/RFP values) ± SD from triplicate experiments. Data are representative of three independent experiments. Source data are provided as a Source Data file.