Figure.
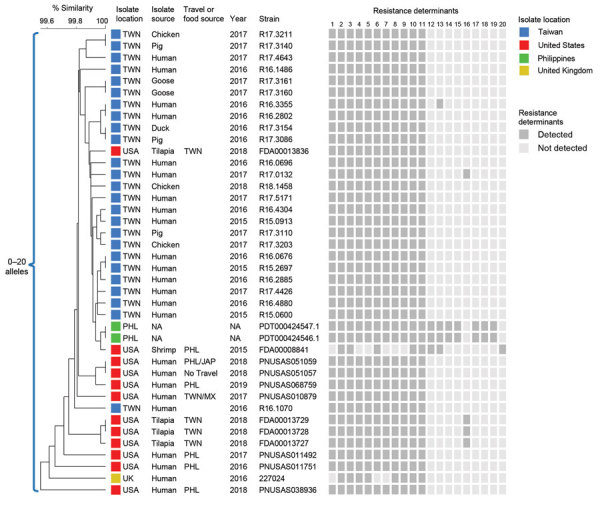
Core genome multilocus sequence typing (cgMLST) phylogenetic tree of 40 Salmonella enterica serotype Anatum isolates, 2015–2019. The tree was constructed by using BioNumerics version 7.6 (Applied Maths, http://www.applied-maths.com). Isolate sources, collection years, and National Center for Biotechnology Information strain or isolate numbers are shown. For isolates from the United States, international travel destinations of patients and sources of imported foods are provided. Dark gray boxes indicate resistance determinants detected: 1) aadA2; 2) aph(3″)-Ib (strA); 3) aph (6)-Id (strB); 4) blaDHA-1; 5) dfrA23; 6) floR; 7) lnu(F); 8) qnrB4; 9) sul1; 10) sul2; 11) tet(A); 12) aadA1; 13) blaTEM-1B; 14) dfrA1; 15) dfrA12; 16) mcr-1.1; 17) mph(A); 18) oqxAB; 19) qnrA6; 20) sul3. Scale bar indicates percentage similarity. JAP, Japan; MX, Mexico; NA, not available; PHL, Philippines; TWN, Taiwan; UK, United Kingdom; USA, United States.
