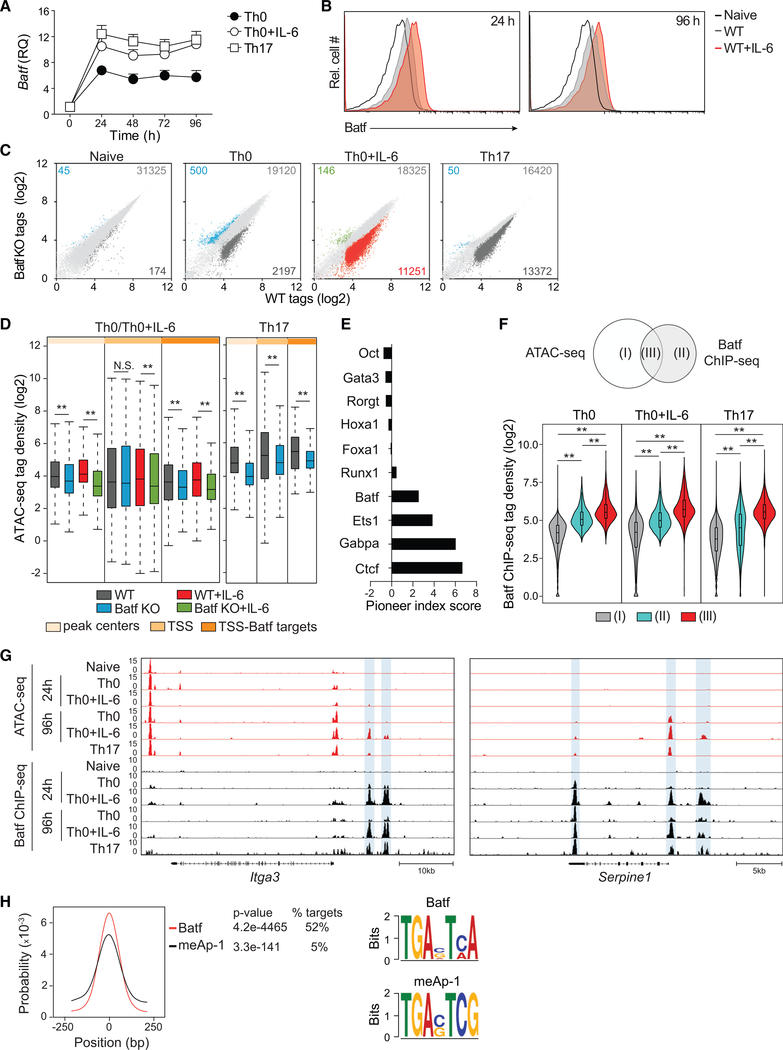
Figure 1. Batf Is Required for Chromatin Accessibility in CD4 + T Cells.
(A) qRT-PCR analysis of Batf expression in naive WT CD4+CD62hi T cells activated for the indicated time points with anti-CD3 and anti-CD28 in the presence or absence of IL-6 or under Th17 culture conditions. Results are relative to naive cell expression (day 0).
(B) Naive WT CD4+CD44lowCD62Lhi T cells were activated with anti-CD3 and anti-CD28, with or without IL-6, for 24 h and 96 h. Cells were harvested and stained intracellularly for Batf.
(C and D) Differential genome-wide chromatin accessibility in WT versus Batf-deficient T cells. Nuclei isolated from naive, 24-h-activated CD4+ T cells ± IL-6, and 48-h Th17 cells from WT and Batf KO cells were subjected to ATAC-seq. Scatterplots of normalized tag density identify chromatin accessibility in WT compared with Batf KO cells (fold change > 2; false discovery rate [FDR] < 0.05) in naive, Th0, Th0+IL-6, and Th17 cells (C). Boxplots indicate normalized signal density ± 500 bp of peak centers (light orange), transcription start sites (TSSs; medium orange), and TSS of Batf target genes (dark orange) that compare WT and Batf KO cultured cells (D).
(E) Factors with pioneering characteristics are enriched in Th0+IL-6 cells. ATAC-seq data from 24-h IL-6-stimulated CD4+ T cells interrogated with the Protein Interaction Quantitation algorithm (PIQ) identified enriched motifs. Greater pioneer index scores correlate with greater chromatin opening.
(F–H) Venn diagram displaying overlapping regions of Batf ChIP-seq and ATAC-seq peaks (left). Violin plots show normalized tag density of Batf binding in naive, Th0 (24 h), Th0+IL-6 (24 h), and Th17 (48 h) cells centered ± 500 bp of unique and overlapping sites between Batf-occupied sites and accessible regions (F). ATAC-seq and Batf ChIP-seq tracks visualized with the Integrated Genome Browser (IGB) identify open chromatin (red) and Batf binding (black) in the Itga3 and Serpine1 genomic regions. Blue highlights indicate Batf binding in closed chromatin regions (G). Batf and meAP-1 motif-enrichment analysis of pooled Batf-bound closed chromatin from naive, 24-h activated CD4+ T cells ± IL-6 and 48-h Th17 cells (equivalent to cluster II in Figure 1F) (H).
Data are means ± SEM of three to five independent experiments with one mouse per experiment (A and B), or representative of two independent experiments with similar results (C–H). RQ, relative quantification; N.S., not significant. **p < 0.001 (Mann-Whitney test).