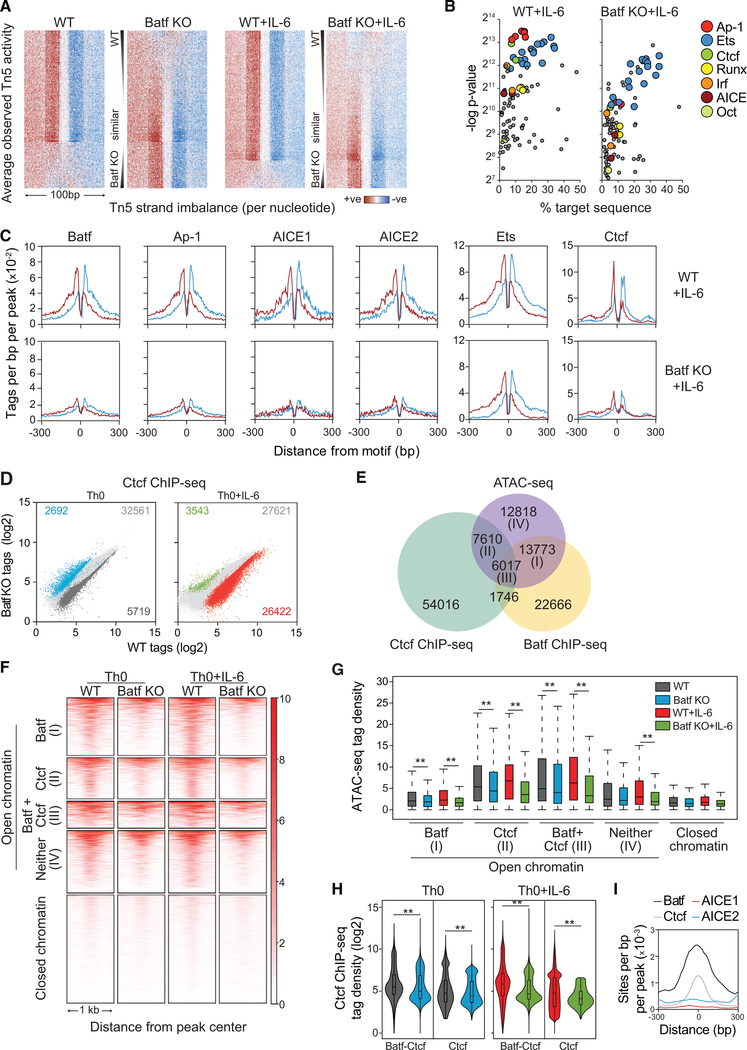
Figure 4. Batf-Mediated Chromatin Accessibility Is Required for Transcription Factor Recruitment in Activated CD4+ T Cells.
(A–C) ATAC-seq data from WT and Batf KO CD4+ T cells activated for 24 h ± IL-6 were subjected to Wellington footprinting analysis (Figures 1C and 1D).Heatmaps show differential footprints compared between WT and Batf KO cells; red indicates positive strand cuts (+ve) over negative strand cuts (ve) per nucleotide positions, and blue indicates negative strand cuts. Binding sites are sorted from top to bottom in order of decreased footprint occupancy score (A). Footprints from WT and Batf KO Th0+IL-6 cells (A) were subjected to motif-enrichment analysis and are represented by scatterplots (B). Histograms show specific transcription factor motif enrichment from footprinting analysis (A), with red indicating positive strand cuts and blue indicating negative strand cuts in WT and Batf KO Th0+IL-6 cells (C).
(D) Scatterplots show normalized Ctcf ChIP-seq tag density with numbers of differential peaks (fold change > 2; FDR < 0.05) from WT and Batf KO Th0 andTh0+IL-6 cells.
(E) Venn diagram displays shared binding of Batf and Ctcf to accessible chromatin regions identified by Batf and Ctcf ChIP-seq from WT Th0+IL-6 cells peaks andmerged ATAC-seq peaks from WT Th0 and Th0+IL-6 and Batf KO Th0 and Th0 +IL-6 cells using the HOMER function mergePeaks with parameter −d given.
(F and G) Heatmap density (F) and boxplots (G) show normalized ATAC-seq signals centered ± 500 bp of the indicated clusters (E).
(H and I) Violin plots of Ctcf occupancy ± 500 bp of overlapped Batf-Ctcf sites or unique Ctcf sites in 96-h WT and Batf KO Th0 and Th0+IL-6 (H). Enriched Batf, Ctcf, AICE1, and AICE2 motifs at genomic sites co-bound by Batf and Ctcf (I).
Data are representative of two independent experiments with similar results. **p < 0.001 (Mann-Whitney test)