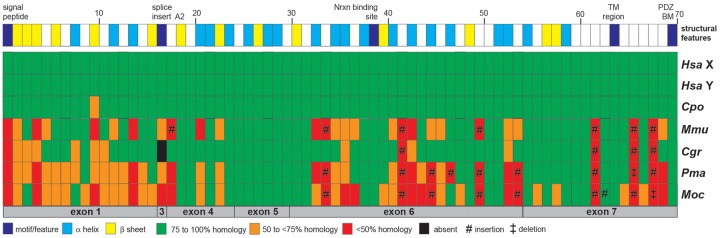
Fig. 4.
Quality of structural differences among mouse and hamster NLGN4 proteins. The NLGN4 protein sequences of human NLGN4X/Y (Hsa X, XP_016885179.1; Hsa Y, XP_011529732.1), guinea pig (Cpo, inferred from its gene, Gene ID: 100714631), mouse (Mmu, CAST/EiJ, MH051930), prairie vole (Moc), Chinese hamster (Cgr), and deer mouse (Pma) (all three hamster NLGN4 protein sequences are included in supplementary text file S1, Supplementary Material online) were aligned and structural features assigned based on the crystal structure information of human NLGN4X (Leone et al. 2010). The alignment was split up into 70 segments. The segments reflected assigned structural features such as the signal peptide, alpha helices, beta strands, the transmembrane region (TM), and the PDZ-binding motif (PDZ BM), but also insertions and low homology regions. Individual segments may vary in size. A detailed annotation of the sequence alignment can be found in Text File S1. The color-code of the heat map reflects the degree of similarity of all species (including human NLGN4Y) relative to the reference sequence of human NLGN4X, which by default is represented entirely by green boxes. Guinea pig NLGN4 was chosen as an example outside the clade eumuroidea due to its high similarity to both human homologs. All segments are displayed relative to their position within the respective exon (gray boxes on the bottom of the heat map). Alignment was performed using MultAlin with default settings (“Identity-0-1”).