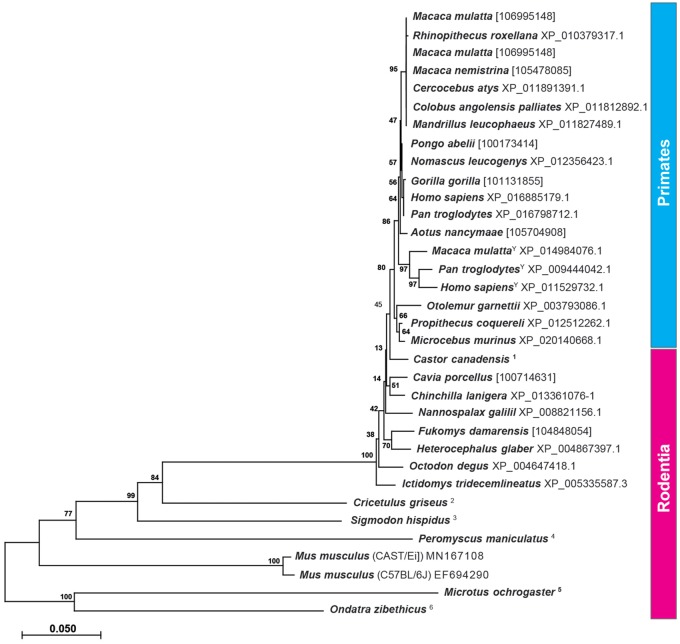
Fig. 5.
Phylogenetic analysis of NLGN4 in primates and rodents. The evolutionary history of neuroligin-4 in primates and rodents based on the translated protein sequences was inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 1.50308039 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 34 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 1,444 positions in the final data set. Evolutionary analyses were conducted in MEGA X. Only protein sequences that included splice insert A2 have been used with the exception of Cricetulus griseus and Ondatra zibethicus in which splice site A2 (exon 3) is absent. In other species, which lacked available protein sequences including splice insert A2, exon 3 had been identified from the respective gene and translated into the final sequences. In these cases, the Gene ID is provided instead. For the primate species, Macaca mulatta, Pan troglodytes, and Homo sapiens, available NLGN4X and NLGN4Y sequences were included. Y, NLGN4Y protein sequence. For the following species, protein sequences were inferred from available genomic DNA: 1, LOC109703453, LOC109674687, and LOC109679926; 2, NW_006882737.1, FYBK01068127.1, FYBK01060804.1, FYBK01060803.1; 3, PVIH01022615.1; 4, NW_006501439.1, MN172170, MN379652; 5, NW_004949307.1, MN841979; 6, PVIU01025695.1.