Fig. 6.
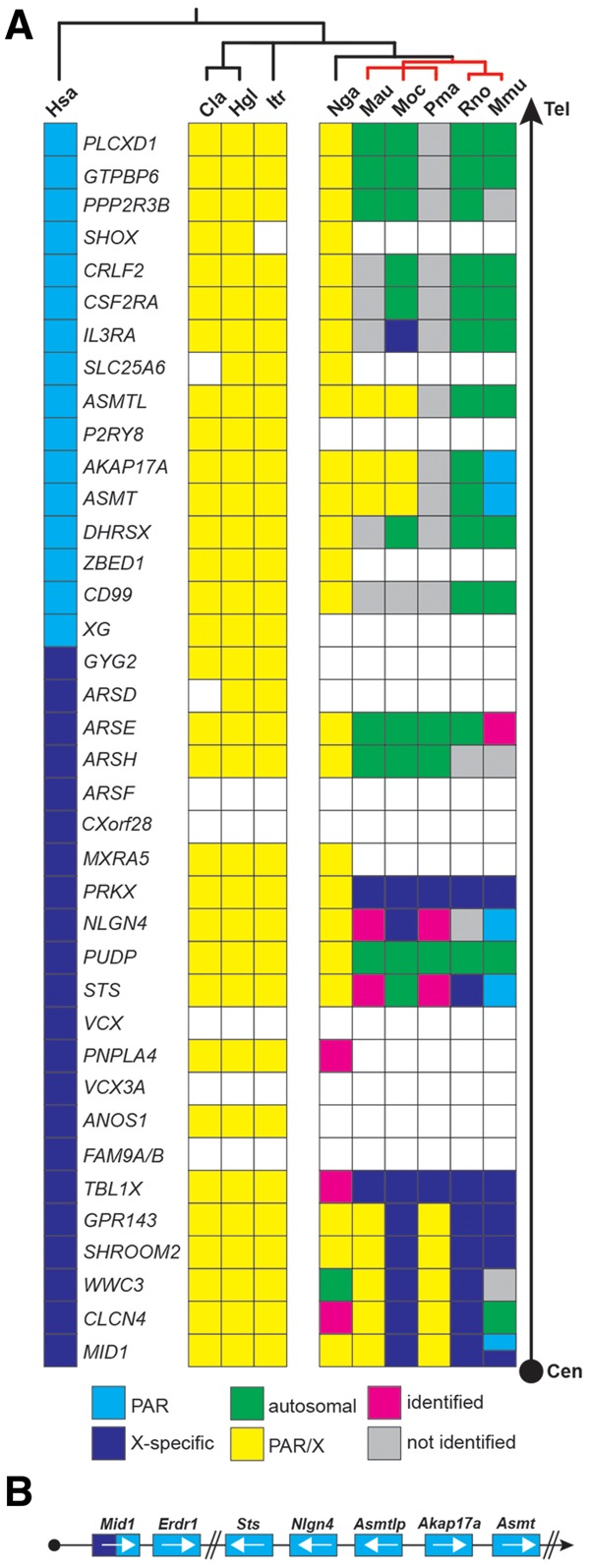
The PAR genes in rodents. (A) Schematic of the localization of the genes of the X-specific and pseudoautosomal region (PAR) of the human X-chromosome compared with selected rodents (for full annotation, see supplementary fig. S3, Supplementary Material online). All genes are listed starting with MID1, a gene that is known to harbor the PAR boundary in mice, to the most telomeric gene in humans, PLCXD1 (Tel, telomere; Cen, direction to centromere). The human PAR boundary, in contrast, is located between GYG2 and XG. As the PAR boundary in most rodents has not been determined yet, the genes are depicted in yellow without formal assignment to either X-specific (indigo) or PAR-specific regions (blue). Genes that are missing within an available contig are shown as open boxes indicating their possible absence from the respective PAR. Genes that have not been identified yet but are likely present in accordance with the overall evolutionary genomic comparison are shown as gray boxes. In case of translocation from pseudoautosomal genes to autosomes the genes are shown in green. Magenta boxes represent genes that were identified in the respective genomes but could not be clearly assigned due to the lack of flanking sequence information. Red bars represent genomes that belong to the eumuroidea clade. Cla, Chinchilla lanigera; Hgl, Heterocephalus glaber; Hsa, Homo sapiens; Itr, Ictidomys tridecemlineatus; Mau, Mesocricetus auratus; Mmu, Mus musculus; Moc, Microtus ochrogaster; Nga, Nannospalax galili; Pma, Peromyscus maniculatus; Rno, Rattus norvegicus. (B) Relative position of the PAR genes (not drawn to scale) in mouse including the mouse-specific Erdr1 gene downstream of Mid1 and the Asmtl pseudogene, Asmtlp. White arrows indicate the orientation of the respective genes on the contig and in the PAR.
