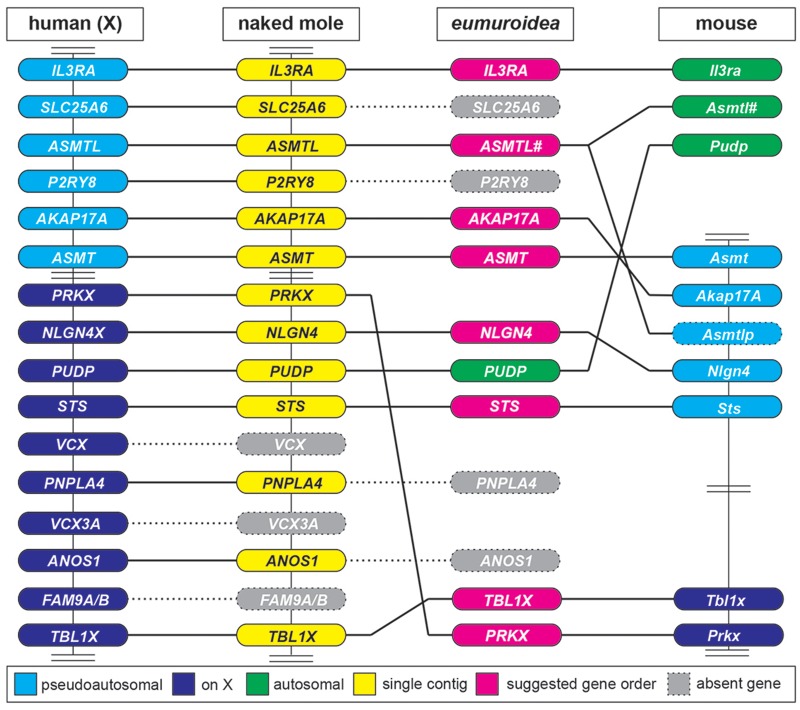
Fig. 7.
Major rearrangements within the PAR and X-chromosome of the clade eumuroidea. This is a horizontal comparison of selected genes from the human X-chromosome (on the left side, including X-specific and PAR genes) with orthologs found in the naked mole (Heterocephalus glaber) and mouse genomes. The naked mole was chosen because all depicted genes in this particular species are found to localize on a single contig (NW_004624834.1). Several critical rearrangements are suggested at the node of the eumuroidea diversification, which itself is depicted as a hypothesized intermediate. VCX and FAM9A/B genes, both found on the human X-chromosome, are absent in rodents, further loss of ANOS1, PNPLA4, P2RY8, and SLC25A6 genes occurs at the node of the eumuroidea branch. The loss of SLC25A6 leads to the truncation of ASMTL. The distantly located genes TBL1X and PRKX recombine within the X-chromosome and have become neighboring genes. PUDP translocates to an autosome allowing STS to join NLGN4. In mice, Il3ra and Asmtl have translocated to different autosomes leaving a copy of Asmtl behind that has subsequently diverged into the pseudogene Asmtlp in the PAR of laboratory mice. Vertical bars connecting genes indicate that based on sequence information all genes have been shown to be present on one strand. Double bars represent gaps that exclude additional genes that are of no immediate relevance in this comparison. #, truncated version of ASMTL upon genomic recombination.