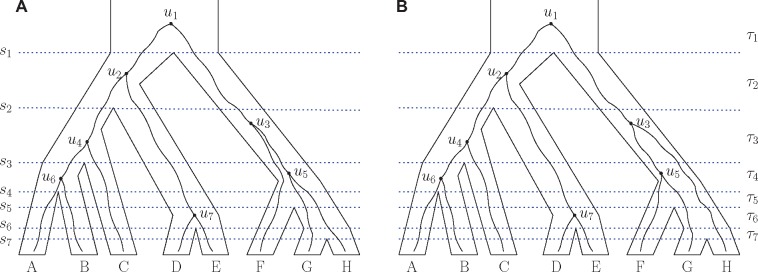
Fig. 10.
Gene trees evolving on an eight-taxon species tree. (A) Ranked gene tree that shares the same unranked topology with that of the species tree. (B) Gene tree that has a different unranked topology from the species tree. Note that the ranked gene tree (not shown) has exactly the same probability as gene trees in (A) and (B) for the species tree depicted. For each denotes the time of the ith speciation, τi represents the interval between the th and ith speciation events, ti () represents the length of interval τi, and ui represents the ith coalescence (node with rank i) in the gene tree. The species tree has ranked topology . For the species tree values , the ranked gene trees in (A) and (B) are the most probable ranked gene trees, with probability .