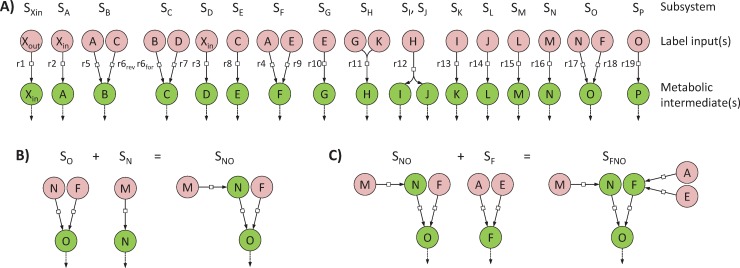
Fig 2. Network decomposition to construct flux models.
The metabolic network shown in Fig 1A can be decomposed into 17 minimal subsystems (panel A) which are sufficient to simulate the labeling dynamics of metabolic intermediates (green circles) from the local label input(s) (red circles). Each minimal subsystem is self-consistent and can be used for independent flux calculations. These minimal subsystems can also be combined to analyze larger subsystems, as shown in panels B and C.