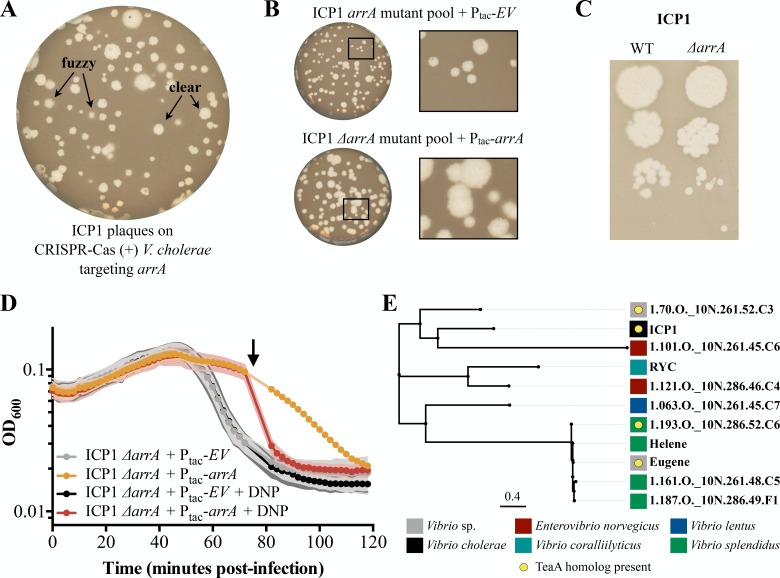
Figure 3. Antiholin ArrA identification and characterization.
(A) Initial plaquing of wild type ICP1 on CRISPR-Cas (+) V. cholerae targeting arrA yielded a mixture of plaque phenotypes. (B) Plaques with clear edges can be found in populations of phage that overcame targeting with various mutations in arrA when plaqued on V. cholerae harboring an empty vector (EV) control (Top). Providing ArrA in trans restores the fuzzy edge phenotypes within the mutant population. (C) Using a repair template we created a clean arrA deletion and found that ΔarrA ICP1 yield plaques with clear edges. (D) During infection, once lysis begins as observed through changes in optical density (OD600), ΔarrA ICP1 causes rapid lysis consistent with abolishment of lysis inhibition. When ArrA is expressed in trans the lysis inhibition phenotype is rescued: a small lysis event is visible 40 minutes post-infection after which the optical density is stabilized for about 30 minutes. Consistent with ArrA restoring lysis inhibition, the stabilization of optical density is sensitive to chemical disruption of the proton motive force by 2,4-dinitrophenol (DNP) (arrow). Points show the average of four or greater replicates; shading shows the standard deviation. (E) BLASTP was used to find proteins with 20% identity to ArrA over 75% of the query; these homologs are displayed in an unrooted tree displaying the name of the phage the protein is found in. Colored blocks show the identity of the host that each phage infects, and yellow circles denote that a protein with homology to TeaA is present in the phage. A table with extended information for each homolog is available in Supplementary file 3.