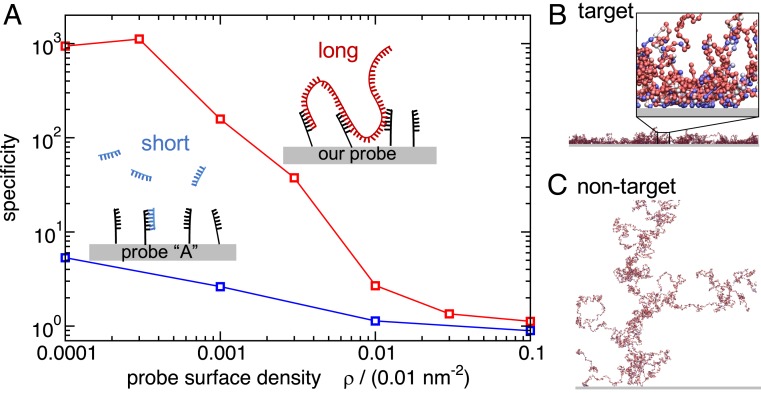
Fig. 1.
Simulating multivalent detection of bacterial DNA. (A) Simulation results for the specificity of binding of target vs. nontarget DNA to surfaces coated with probes targeting E. coli. Specificity is defined as the ratio of DNA–surface contacts for the target E. coli versus nontarget DNA from the bacterium B. subtilis. The blue curve shows results for a published 40-nt probe, “probe A” (18) that targets the 16S ribosomal gene of E. coli, binding to DNA fragmented into 400-nt segments. The red curve shows results for our top-scoring multivalent 20-nt probe binding to unfragmented genomic DNA (for the same total amount of DNA as in the blue curve). (B and C) Snapshots from our simulations for genomic DNA of E. coli (B) and B. subtilis (C) binding to the surface coated in multivalent probes. The genomic DNA is modeled as a chain of 400-nt “blobs.” Each ball represents a single blob and its color indicates the interaction strength between the blob and the surface: weak interaction, red; strong interaction, blue. The blow-up in B, Inset shows that blue blobs with a stronger surface-binding interaction are predominantly found close to the surface. The density of probes on the surface was set to . The lateral size of the snapshots is m, while the Inset to B is of size 300 nm.