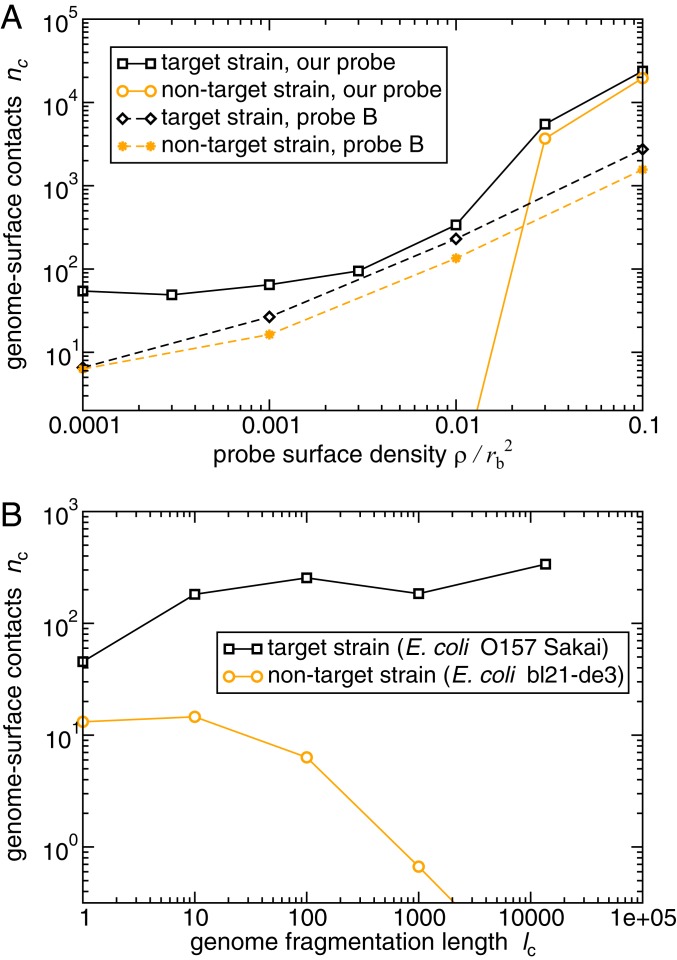
Fig. 3.
Distinguishing between similar genomes: the target strain E. coli O157 Sakai and the nontarget strain E. coli bl21-de3 (wild type). (A) Results of our Langevin dynamics simulations for the average degree of binding (number of blob–surface contacts ), for the target and nontarget genomic DNA. The solid lines show results for the top-scoring 20-nt probe, which is designed to maximize the difference function . The dashed lines show results for the published 70-nt “probe B” (38), which targets the rfbE gene specific to E. coli O157. For the published probe, the genomic DNA has been fragmented into 400-nt pieces to mimic standard DNA microarray conditions while for our 20-nt probe we assume unfragmented genomic DNA. (B) The effect of genome fragmentation at surface probe density , for our top-scoring 20-nt probe. As in our other simulations, , and a genome–surface contact was defined as a DNA blob being located within 2 nm of the surface.