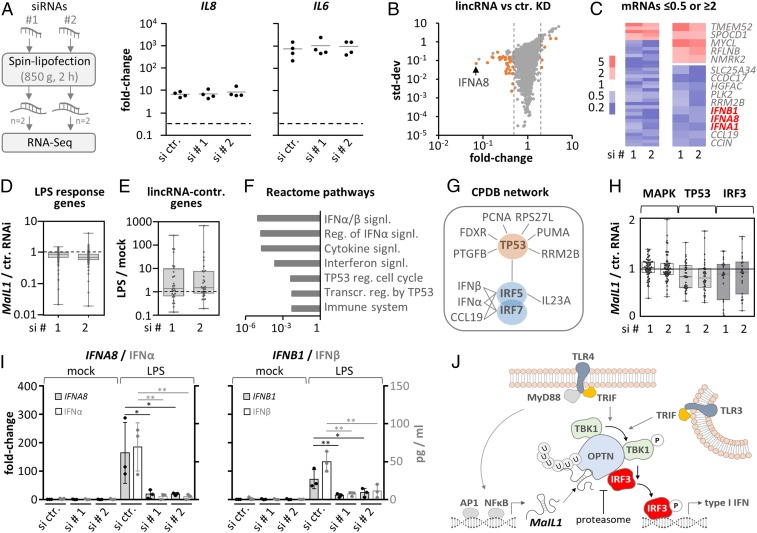
Fig. 6.
MaIL1 controls type I IFN expression. (A, Left) Knockdown strategy using independent siRNA designs. (A, Right) qRT-PCR analysis of IL8 and IL6 induction (8-h LPS versus mock, four replicates), in MaIL1-silenced macrophages. (B) RNA-seq plot showing mean fold changes and SD (MaIL1 siRNA 1 or 2 vs. control siRNA knockdown). Genes with fold changes ≥2 or ≤0.5 in both runs are highlighted (orange). (C) Fold change heatmap for orange-labeled genes from B. Five up- and top 10 down-regulated genes are shown to the Right. (D) Box plot showing regulation of LPS-response genes (RNA-seq determined, fold change ≥ 2, LPS vs. mock) upon MaIL1 compared to control knockdown. (E) Box plot showing RNA-seq–determined regulation of MaIL1-controlled genes from C in response to LPS. (F) Reactome pathway analysis using down-regulated genes from C. (G) ConsensusPathDB network analysis using down-regulated genes from C. (H) Regulation of MAPK-, TP53-, and IRF3-dependent genes (MaIL1 versus control knockdown, data from B). (I) qRT-PCR and ELISA validation of diminished type I IFN mRNA (primary axis) and protein (secondary axis) induction upon MaIL1 knockdown (mRNA fold changes compared to control siRNA and mock stimulation, three replicates). (J) Model: MaIL1 fosters OPTN aggregation to promote TLR-TRIF-TBK1–dependent IRF3 phosphorylation and type I IFN expression. MaIL1 expression and thereby TLR–TRIF activity is induced by TLR–Myd88 signaling. The proteasome eventually resolves MaIL1-dependent OPTN aggregates. *P ≤ 0.05, **P ≤ 0.01 (one-way ANOVA).