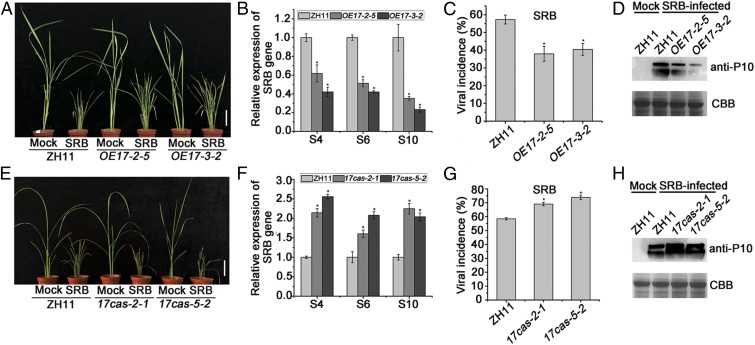
Fig. 4.
OsARF17 positively modulates rice resistance to SRBSDV. (A and E) The symptoms on SRBSDV-infected WT (ZH11), transgenic, and mutant plants. The phenotypes were observed and photos taken at 30 dpi. (Scale bars, 5 cm.) (B and F) qRT-PCR results showing the relative expression levels of SRBSDV (S4, S6, and S10) in SRBSDV-infected OE17 (OE17-2-5 and OE17-3-–2) and 17cas (17cas-2-1 and 17cas-5-2) plants compared with SRBSDV-infected control plants. UBQ5 was used as the internal reference gene to normalize the relative expression. Values shown are the means ± SD of 3 biological replicates. Significant differences were identified using Fisher's least significant difference tests. *At the top of columns indicates significant difference at P ≤ 0.05. (C and G) Disease incidence in ZH11 (control) and OE17 lines (OE17-2-5 and OE17-3-2) or 17cas (17cas-2-1 and 17cas-5-2) lines, respectively. The percentage of plants infected by SRBSDV was determined by RT-PCR at 30 dpi. (D and H) The accumulation of SRBSDV P10 protein in SRBSDV-infected plants determined by Western blotting.