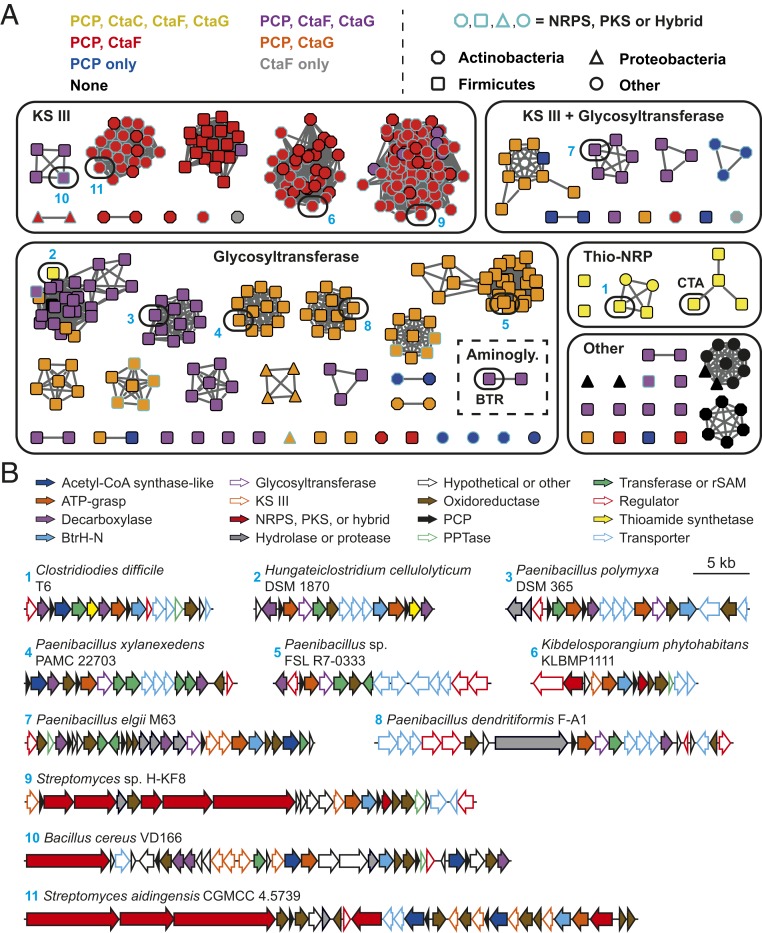
Fig. 5.
Genome mining for additional noncanonical thiotemplate pathways. (A) Sequence similarity network for CtaD homologs (95% identical sequences are conflated; alignment score 115). Nodes are color-coded based on the presence of genes encoding PCPs and homologs of the indicated CTA biosynthetic enzymes in the genomic region surrounding the CtaD homolog. Node shapes correspond to the phylum of the organism harboring the biosynthetic gene cluster, while the color of the node outline indicates the presence of genes encoding an NRPS or PKS in the gene neighborhood. The ATP-grasp proteins from the CTA and butirosin (BTR) biosynthetic pathways are indicated. SI Appendix, Table S4 lists all the sequences in the network. Aminogly., aminoglycoside; Thio-NRP, thioamidated NRP. (B) Representative biosynthetic gene clusters that encode a CtaD homolog (selected based on architecture diversity). Numbers correspond to the node numbering in A. Genes encoding methyltransferases, acetyltransferases, nucleotidyltransferases, aminotransferases, and radical S-adenosyl-l-methionine enzymes are grouped under the designation “Transferase or rSAM”.