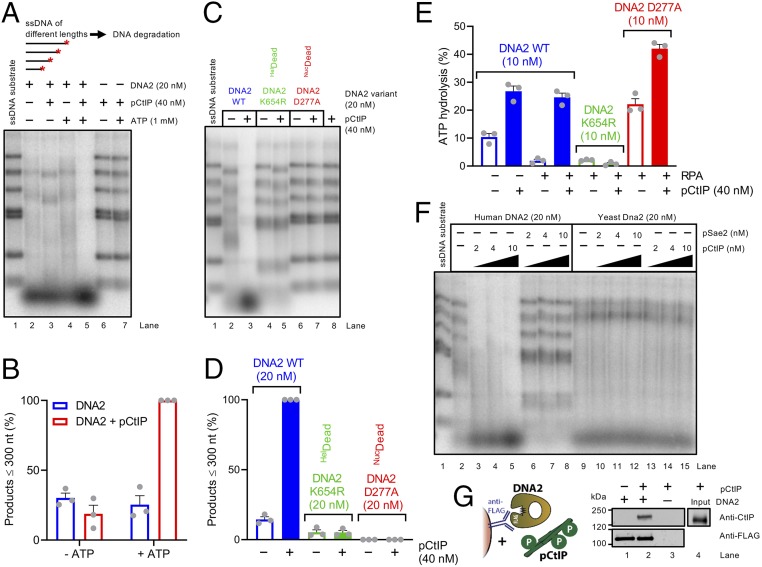
Fig. 3.
The motor activity of DNA2 mediates the accelerated ssDNA degradation with pCtIP. (A) Degradation of 3′ end-labeled ssDNA fragments (derived from λDNA) of various lengths by DNA2 in the presence or absence of pCtIP without or with ATP, as indicated. All reactions contained human RPA (864 nM). The experiment was incubated at 37 °C for 8 min. ATP is required for the stimulatory effect of pCtIP on DNA2. A red asterisk indicates the position of the labeling. (B) Quantitation of data such as shown in A. n = 3; error bars, SEM. The grey circles represent data points from indepedent experiments. (C) Degradation of ssDNA fragments by WT, helicase-deficient K654R, or nuclease-deficient D277A DNA2 variants without or with pCtIP. All reactions contained human RPA (864 nM). The experiment was incubated at 37 °C for 8 min. HelDead, helicase-dead; NucDead, nuclease-dead. (D) Quantitation of small degradation products from experiments such as shown in C. n = 3; error bars, SEM. (E) ATP hydrolysis by WT, helicase-deficient K654R, or nuclease-deficient D277A DNA2 alone (10 nM) or with pCtIP (40 nM). Reactions contained 10.3-kbp-long substrate denatured at 95 °C for 5 min, 395.5 nM human RPA where indicated and 20 mM NaCl. (F) Degradation of 3′ end-labeled ssDNA fragments derived from λDNA by human or yeast DNA2/Dna2 without or with human pCtIP or yeast pSae2. Reactions with human DNA2 were carried out at 37 °C for 8 min with human RPA (864 nM). Reactions with yeast Dna2 were performed at 25 °C for 1 min with yeast RPA (1.09 µM). (G) Analysis of DNA2 interaction with pCtIP. DNA2-FLAG was immobilized on M2 anti-FLAG affinity resin and incubated with purified recombinant pCtIP (phosphorylation sites depicted as P in green circles). The Western blot was performed with anti-FLAG and anti-CtIP antibodies.