Figure 2. The structure of γTuSC.
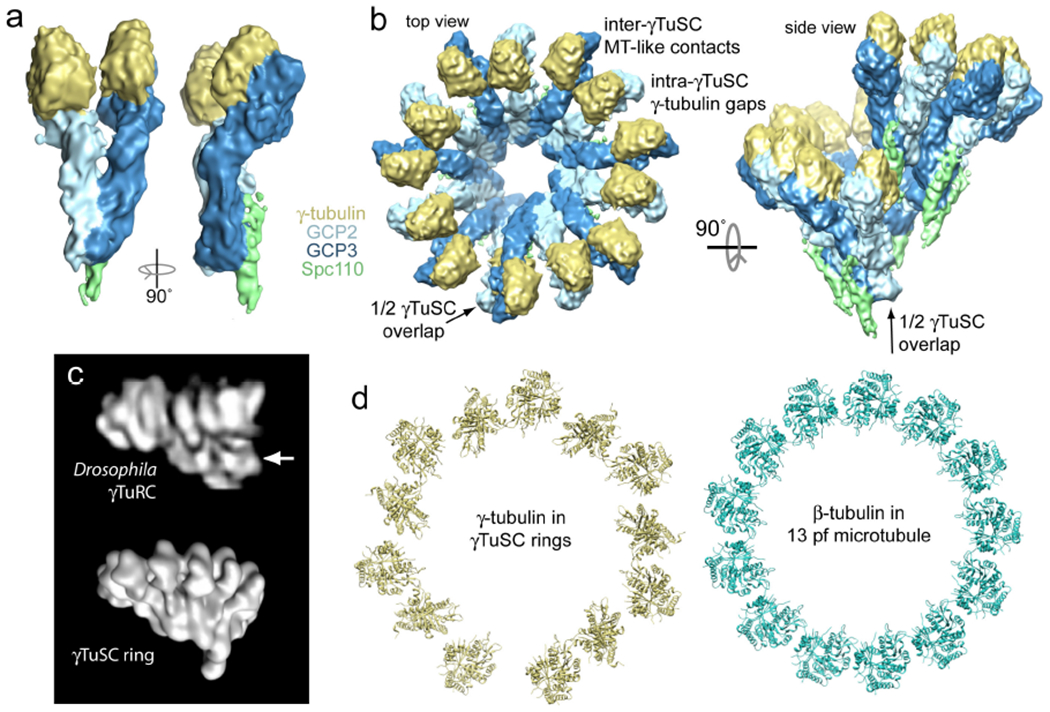
a) The 8 Å cryo-electron microscopy (EM) structure of S. cerevisiae γTuSC bound to the attachment factor Spc110 is shown. This γTuSC is a single subunit of a large γTuSC oligomer (see panel b). In this view, the N-termini of GCP2 and GCP3 are at bottom, with their C-terminal domains near the top interacting with γ-tubulin. In the structure, the two γ-tubulins are held separated from each other in a configuration incompatible with the microtubule lattice, which partially explains the relatively low nucleating capacity of free γTuSC relative to γTuRC. b) Top-down and side views of the γTuSC ring are shown. The ring has six and a half γTuSCs per turn, which arise due to a half γTuSC overlap between the first and seventh subunits in the ring (see side view). This yields thirteen γ-tubulins per turn, matching the in vivo microtubule protofilament number. The conformation of γTuSC is unchanged in the ring structure, such that the intra-γTuSC gap between γ-tubulins remains. However, microtubule-like lateral interactions are observed between γ-tubulins at the inter-γTuSC interface (Ref. 49). c) The low-resolution negative stain EM reconstruction of a single Drosophila melanogaster γTuRC (top, Ref. 40) closely resembles the γTuSC ring shown in panel b, rendered here at lower resolution for comparison (bottom). The region of γTuRC originally interpreted as a GCP4,5,6 cap is indicated with an arrow; this region appears to correspond to the N-terminal regions of GCP2 and GCP3 instead. d) Comparison of γ-tubulin positions in γTuSC rings and αβ-tubulin in the microtubule shows a mismatch in geometry, with alternating contacts and gaps in the γ-tubulin arrangement (Ref. 49).
