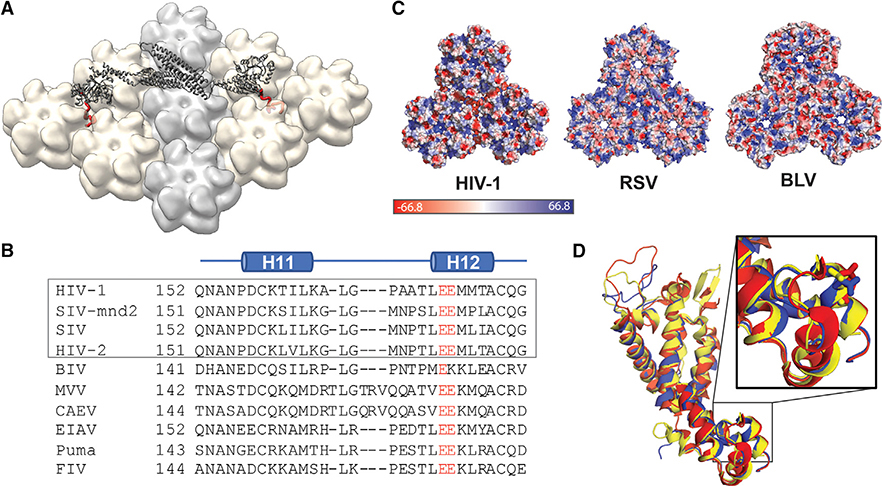
Figure 5. MxB Recognizes CA Features that Are Potentially Conserved in Lentiviruses but not in Retroviruses.
(A) Model of MxB binding to the CA lattice, generated using existing structural data of the MxB dimer (gray, PDB: 4WHJ) and HIV CA (light gray/tan, PDB: 4XFX) and MD simulation data of the MxB N-terminal peptide binding to CA (red).
(B) Alignment of CA structures available from different lentiviruses: HIV (PDB: 4XFX, red), equine infectious anemia virus (EIAV) (PDB: 2EIA, blue), and feline immunodeficiency virus (FIV) (PDB: 5NA2, yellow); residue E213 shown in sticks located in the same position in each CA.
(C) CA protein sequence alignment of lentiviruses. Gray box denotes primate lentiviruses, which are restricted by MxB. Residue E213 and its homologous residues (red) are conserved in the lentiviral family.
(D) Surface electrostatic potential distributions of diverse retroviral capsids show few shared characteristics at the tri-hexamer interface. The following structures are used in comparison: HIV-1 (PDB: 4XFX), Rous sarcoma virus (RSV) (PDB: 4TIR), and bovine leukemia virus (BLV) (PDB: 4PH0).