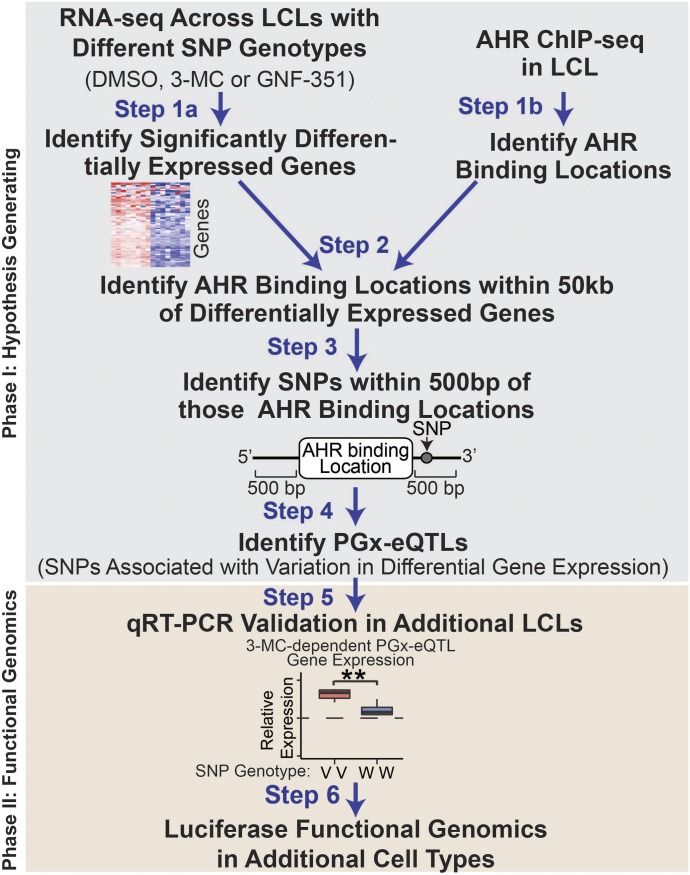
Fig. 1.
PGx-eQTL analysis scheme. A PGx pipeline was developed to merge RNA-seq, ChIP-seq, and SNP-genotype data to identify putative PGx-eQTLs. As a first step, we identified the top differentially expressed genes [|Log2(fold change)| >1 and FDR <0.05] from our RNA-seq analyses for eight LCLs that were treated with 0.1% DMSO, 1 µM 3-MC, or 1 µM GNF-351 in duplicate (Step 1a). In parallel, we identified AHR-binding regions from AHR ChIP-seq analysis performed for one of the LCLs used in the RNA-seq studies (Step 1b). We next determined which of the top differentially expressed genes had AHR-binding peaks within 50 kb of the gene (Step 2) and identified those peaks that had SNPs with a MAF >0.25 that mapped within 500 bp of AHR ChIP-seq peaks (Step 3). We then identified putative PGx-eQTLs by testing the association of the SNPs from Step 3 with the differential expression of the nearby differentially expressed gene with the Spearman rank correlation and tested for significance under a two-tailed t-distribution (Step 4). We validated the putative PGx-eQTLs identified in additional LCLs (Step 5) and functionally validated them with luciferase reporter gene assays (Step 6).