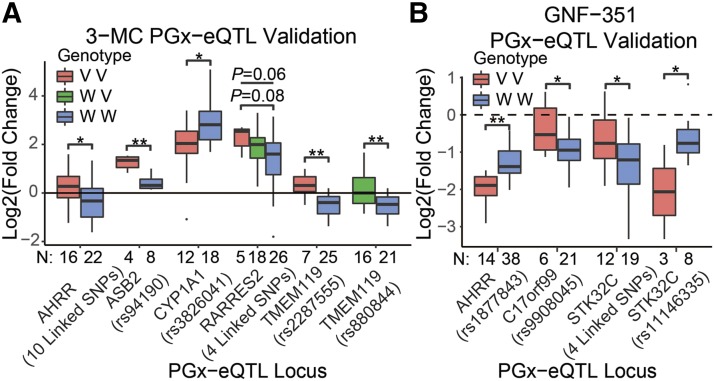
Fig. 3.
Validation of PGx-eQTL. Nine of the 15 testable putative PGx-eQTLs were validated by qRT-PCR, and one was suggestive. (A) Six validated PGx-eQTL loci were 3-MC dependent and (B) four validated PGx-eQTLs were GNF-351 treatment dependent. None of the loci were both 3-MC and GNF-351 treatment dependent. When possible, cell lines that were either homozygous variant (red) or homozygous wild-type (blue) were used to test differential expression. When unavailable (TMEM119-rs880844) or when a suggestive P value was detected (RARRES2-4–linked SNPs), cell lines heterozygous for those SNPs were used. All tests for significance used a two-tailed Student’s t test, and ANOVA was used across the three genotypes for the RARRES2 locus. The number of samples (N) for each group is at the bottom of the x-axis for each plot. *P < 0.05; **P < 0.01.